Circadian rhythms. A protein fold switch joins the circadian oscillator to clock output in cyanobacteria
- PMID: 26113641
- PMCID: PMC4506712
- DOI: 10.1126/science.1260031
Circadian rhythms. A protein fold switch joins the circadian oscillator to clock output in cyanobacteria
Abstract
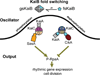
Organisms are adapted to the relentless cycles of day and night, because they evolved timekeeping systems called circadian clocks, which regulate biological activities with ~24-hour rhythms. The clock of cyanobacteria is driven by a three-protein oscillator composed of KaiA, KaiB, and KaiC, which together generate a circadian rhythm of KaiC phosphorylation. We show that KaiB flips between two distinct three-dimensional folds, and its rare transition to an active state provides a time delay that is required to match the timing of the oscillator to that of Earth's rotation. Once KaiB switches folds, it binds phosphorylated KaiC and captures KaiA, which initiates a phase transition of the circadian cycle, and it regulates components of the clock-output pathway, which provides the link that joins the timekeeping and signaling functions of the oscillator.
Copyright © 2015, American Association for the Advancement of Science.
Figures



References
-
- Dunlap JC. Molecular basis for circadian clocks. Cell. 1999;96:271–290. - PubMed
-
- Nakajima M, et al. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science. 2005;308:414–415. - PubMed
-
- Ishiura M, et al. Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science. 1998;281:1519–1523. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

