Gut resistome development in healthy twin pairs in the first year of life
- PMID: 26113976
- PMCID: PMC4480905
- DOI: 10.1186/s40168-015-0090-9
Gut resistome development in healthy twin pairs in the first year of life
Erratum in
-
Erratum: Gut resistome development in healthy twin pairs in the first year of life.Microbiome. 2015 Jul 23;3:29. doi: 10.1186/s40168-015-0095-4. eCollection 2015. Microbiome. 2015. PMID: 26207183 Free PMC article.
Abstract
Background: The early life of the human host marks a critically important time for establishment of the gut microbial community, yet the developmental trajectory of gut community-encoded resistance genes (resistome) is unknown. We present a longitudinal study of the fecal antibiotic resistome of healthy amoxicillin-exposed and antibiotic-naive twins and their mothers during the first year of life.
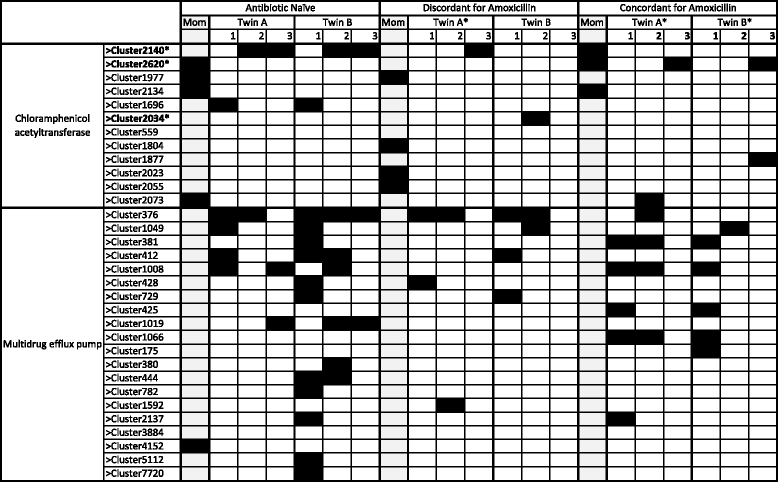
Results: We extracted metagenomic DNA (mgDNA) from fecal samples collected from three healthy twin pairs at three timepoints (1 or 2 months, 6 or 7 months, and 11 months) and from their mothers (collected at delivery). The mgDNA was used to construct metagenomic expression libraries in an Escherichia coli host. These libraries were screened for antibiotic resistance, and functionally selected resistance genes were sequenced and annotated. A diverse fecal resistome distinct from the maternal resistome was apparent by 2 months of age, and infants' fecal resistomes included resistance to clinically important broad-spectrum beta-lactam antibiotics (e.g., piperacillin-tazobactam, aztreonam, cefepime) not found in their mothers. Dissemination of resistance genes among members of a given family was positively correlated with sharing of those same resistance genes between unrelated families, potentially identifying within-family sharing as a marker of resistance genes emerging in the human community at large. Finally, we found a distinct developmental trajectory for a community-encoded function: chloramphenicol resistance. All study subjects at all timepoints harbored chloramphenicol resistance determinants, but multidrug efflux pumps (rarely found in mothers) were the primary effectors of chloramphenicol resistance in young infants. Chloramphenicol acetyltransferases were more common in mothers than in infants and were found in nearly all the infants at later timepoints.
Conclusions: Our results suggest that healthy 1-2-month-old infants' gut microbes harbor clinically relevant resistance genes distinct from those of their mothers, and that family-specific shared environmental factors early in life shape resistome development.
Keywords: Antibiotic resistance; Beta lactamase; Chloramphenicol resistance; Fecal microbiome; Gut microbiome; Pediatrics.
Figures




References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

