BreakSeek: a breakpoint-based algorithm for full spectral range INDEL detection
- PMID: 26117537
- PMCID: PMC4538813
- DOI: 10.1093/nar/gkv605
BreakSeek: a breakpoint-based algorithm for full spectral range INDEL detection
Abstract
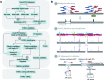
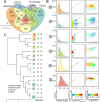
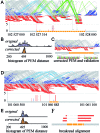
Although recent developed algorithms have integrated multiple signals to improve sensitivity for insertion and deletion (INDEL) detection, they are far from being perfect and still have great limitations in detecting a full size range of INDELs. Here we present BreakSeek, a novel breakpoint-based algorithm, which can unbiasedly and efficiently detect both homozygous and heterozygous INDELs, ranging from several base pairs to over thousands of base pairs, with accurate breakpoint and heterozygosity rate estimations. Comprehensive evaluations on both simulated and real datasets revealed that BreakSeek outperformed other existing methods on both sensitivity and specificity in detecting both small and large INDELs, and uncovered a significant amount of novel INDELs that were missed before. In addition, by incorporating sophisticated statistic models, we for the first time investigated and demonstrated the importance of handling false and conflicting signals for multi-signal integrated methods.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

