Complementing tissue characterization by integrating transcriptome profiling from the Human Protein Atlas and from the FANTOM5 consortium
- PMID: 26117540
- PMCID: PMC4538815
- DOI: 10.1093/nar/gkv608
Complementing tissue characterization by integrating transcriptome profiling from the Human Protein Atlas and from the FANTOM5 consortium
Abstract
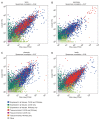
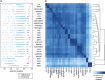
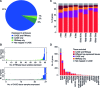
Understanding the normal state of human tissue transcriptome profiles is essential for recognizing tissue disease states and identifying disease markers. Recently, the Human Protein Atlas and the FANTOM5 consortium have each published extensive transcriptome data for human samples using Illumina-sequenced RNA-Seq and Heliscope-sequenced CAGE. Here, we report on the first large-scale complex tissue transcriptome comparison between full-length versus 5'-capped mRNA sequencing data. Overall gene expression correlation was high between the 22 corresponding tissues analyzed (R > 0.8). For genes ubiquitously expressed across all tissues, the two data sets showed high genome-wide correlation (91% agreement), with differences observed for a small number of individual genes indicating the need to update their gene models. Among the identified single-tissue enriched genes, up to 75% showed consensus of 7-fold enrichment in the same tissue in both methods, while another 17% exhibited multiple tissue enrichment and/or high expression variety in the other data set, likely dependent on the cell type proportions included in each tissue sample. Our results show that RNA-Seq and CAGE tissue transcriptome data sets are highly complementary for improving gene model annotations and highlight biological complexities within tissue transcriptomes. Furthermore, integration with image-based protein expression data is highly advantageous for understanding expression specificities for many genes.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Schena M., Shalon D., Davis R.W., Brown P.O. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. - PubMed
-
- Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. - PubMed
-
- Kodzius R., Kojima M., Nishiyori H., Nakamura M., Fukuda S., Tagami M., Sasaki D., Imamura K., Kai C., Harbers M., et al. CAGE: cap analysis of gene expression. Nat. Methods. 2006;3:211–222. - PubMed
-
- Shiraki T., Kondo S., Katayama S., Waki K., Kasukawa T., Kawaji H., Kodzius R., Watahiki A., Nakamura M., Arakawa T., et al. Cap analysis gene expression for high-throughput analysis of transcriptional starting point and identification of promoter usage. Proc. Natl. Acad. Sci. U.S.A. 2003;100:15776–15781. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

