Redefining the transcriptional regulatory dynamics of classically and alternatively activated macrophages by deepCAGE transcriptomics
- PMID: 26117544
- PMCID: PMC4538831
- DOI: 10.1093/nar/gkv646
Redefining the transcriptional regulatory dynamics of classically and alternatively activated macrophages by deepCAGE transcriptomics
Abstract
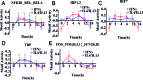
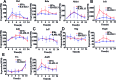
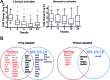
Classically or alternatively activated macrophages (M1 and M2, respectively) play distinct and important roles for microbiocidal activity, regulation of inflammation and tissue homeostasis. Despite this, their transcriptional regulatory dynamics are poorly understood. Using promoter-level expression profiling by non-biased deepCAGE we have studied the transcriptional dynamics of classically and alternatively activated macrophages. Transcription factor (TF) binding motif activity analysis revealed four motifs, NFKB1_REL_RELA, IRF1,2, IRF7 and TBP that are commonly activated but have distinct activity dynamics in M1 and M2 activation. We observe matching changes in the expression profiles of the corresponding TFs and show that only a restricted set of TFs change expression. There is an overall drastic and transient up-regulation in M1 and a weaker and more sustainable up-regulation in M2. Novel TFs, such as Thap6, Maff, (M1) and Hivep1, Nfil3, Prdm1, (M2) among others, were suggested to be involved in the activation processes. Additionally, 52 (M1) and 67 (M2) novel differentially expressed genes and, for the first time, several differentially expressed long non-coding RNA (lncRNA) transcriptome markers were identified. In conclusion, the finding of novel motifs, TFs and protein-coding and lncRNA genes is an important step forward to fully understand the transcriptional machinery of macrophage activation.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
-
- Martinez F.O., Gordon S., Locati M., Mantovani A. Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. J. Immunol. 2006;177:7303–7311. - PubMed
-
- Gordon S. Alternative activation of macrophages. Nat. Rev. Immunol. 2003;3:23–35. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

