Mapping Nucleosome Resolution Chromosome Folding in Yeast by Micro-C
- PMID: 26119342
- PMCID: PMC4509605
- DOI: 10.1016/j.cell.2015.05.048
Mapping Nucleosome Resolution Chromosome Folding in Yeast by Micro-C
Abstract
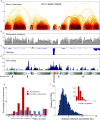
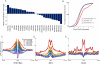
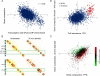
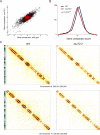
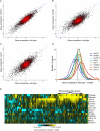
We describe a Hi-C-based method, Micro-C, in which micrococcal nuclease is used instead of restriction enzymes to fragment chromatin, enabling nucleosome resolution chromosome folding maps. Analysis of Micro-C maps for budding yeast reveals abundant self-associating domains similar to those reported in other species, but not previously observed in yeast. These structures, far shorter than topologically associating domains in mammals, typically encompass one to five genes in yeast. Strong boundaries between self-associating domains occur at promoters of highly transcribed genes and regions of rapid histone turnover that are typically bound by the RSC chromatin-remodeling complex. Investigation of chromosome folding in mutants confirms roles for RSC, "gene looping" factor Ssu72, Mediator, H3K56 acetyltransferase Rtt109, and the N-terminal tail of H4 in folding of the yeast genome. This approach provides detailed structural maps of a eukaryotic genome, and our findings provide insights into the machinery underlying chromosome compaction.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures

Comment in
-
Filling the gap: Micro-C accesses the nucleosomal fiber at 100-1000 bp resolution.Genome Biol. 2015 Aug 21;16(1):169. doi: 10.1186/s13059-015-0744-8. Genome Biol. 2015. PMID: 26294274 Free PMC article.
-
Genomics. Micro-C maps of genome structure.Nat Methods. 2015 Sep;12(9):812. doi: 10.1038/nmeth.3575. Nat Methods. 2015. PMID: 26554092 No abstract available.
References
-
- Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science (New York, NY. 2002;295:1306–1311. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

