Structure-Based Design of Inhibitors of Protein-Protein Interactions: Mimicking Peptide Binding Epitopes
- PMID: 26119925
- PMCID: PMC4557054
- DOI: 10.1002/anie.201412070
Structure-Based Design of Inhibitors of Protein-Protein Interactions: Mimicking Peptide Binding Epitopes
Abstract
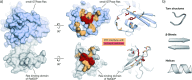
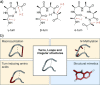
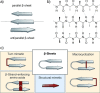
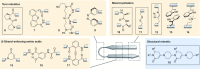
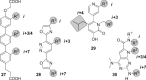
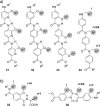
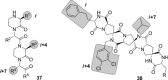
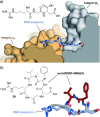
Protein-protein interactions (PPIs) are involved at all levels of cellular organization, thus making the development of PPI inhibitors extremely valuable. The identification of selective inhibitors is challenging because of the shallow and extended nature of PPI interfaces. Inhibitors can be obtained by mimicking peptide binding epitopes in their bioactive conformation. For this purpose, several strategies have been evolved to enable a projection of side chain functionalities in analogy to peptide secondary structures, thereby yielding molecules that are generally referred to as peptidomimetics. Herein, we introduce a new classification of peptidomimetics (classes A-D) that enables a clear assignment of available approaches. Based on this classification, the Review summarizes strategies that have been applied for the structure-based design of PPI inhibitors through stabilizing or mimicking turns, β-sheets, and helices.
Keywords: inhibitors; peptides; peptidomimetics; protein-protein interactions.
© 2015 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.
Figures



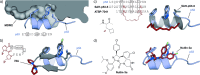

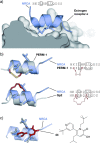

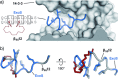
References
-
- Milroy LG, Grossmann TN, Hennig S, Brunsveld L, Ottmann C. Chem. Rev. 2014;114:4695–4748. - PubMed
-
- Moreira IS, Fernandes PA, Ramos MJ. Proteins Struct. Funct. Bioinf. 2007;68:803–812. - PubMed
-
- Geppert T, Hoy B, Wessler S, Schneider G. Chem. Biol. 2011;18:344–353. - PubMed
-
- Huang L, Hofer F, Martin GS, Kim S-H. Nat. Struct. Mol. Biol. 1998;5:422–426. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

