PML is required for telomere stability in non-neoplastic human cells
- PMID: 26119943
- PMCID: PMC4830905
- DOI: 10.1038/onc.2015.246
PML is required for telomere stability in non-neoplastic human cells
Erratum in
-
PML is required for telomere stability in non-neoplastic human cells.Oncogene. 2016 Apr 7;35(14):1876. doi: 10.1038/onc.2015.312. Oncogene. 2016. PMID: 27052595 Free PMC article. No abstract available.
Abstract
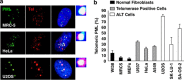
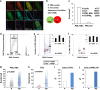
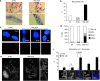
Telomeres interact with numerous proteins, including components of the shelterin complex, whose alteration, similarly to proliferation-induced telomere shortening, initiates cellular senescence. In tumors, telomere length is maintained by Telomerase activity or by the Alternative Lengthening of Telomeres mechanism, whose hallmark is the telomeric localization of the promyelocytic leukemia (PML) protein. Whether PML contributes to telomeres maintenance in normal cells is unknown. We show that in normal human fibroblasts the PML protein associates with few telomeres, preferentially when they are damaged. Proliferation-induced telomere attrition or their damage due to alteration of the shelterin complex enhances the telomeric localization of PML, which is increased in human T-lymphocytes derived from patients genetically deficient in telomerase. In normal fibroblasts, PML depletion induces telomere damage, nuclear and chromosomal abnormalities, and senescence. Expression of the leukemia protein PML/RARα in hematopoietic progenitors displaces PML from telomeres and induces telomere shortening in the bone marrow of pre-leukemic mice. Our work provides a novel view of the physiologic function of PML, which participates in telomeres surveillance in normal cells. Our data further imply that a diminished PML function may contribute to cell senescence, genomic instability, and tumorigenesis.
Figures




References
-
- Akbar AN, Vukmanovic-Stejic M. Telomerase in T lymphocytes: use it and lose it? J Immunol 2007; 178: 6689–6694. - PubMed
-
- d'Adda di Fagagna F, Reaper PM, Clay-Farrace L, Fiegler H, Carr P, Von Zglinicki T et al. A DNA damage checkpoint response in telomere-initiated senescence. Nature 2003; 426: 194–198. - PubMed
-
- Takai H, Smogorzewska A, de Lange T. DNA damage foci at dysfunctional telomeres. Curr Biol 2003; 13: 1549–1556. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

