DNA Electrochemistry Shows DNMT1 Methyltransferase Hyperactivity in Colorectal Tumors
- PMID: 26120002
- PMCID: PMC4518711
- DOI: 10.1016/j.chembiol.2015.05.019
DNA Electrochemistry Shows DNMT1 Methyltransferase Hyperactivity in Colorectal Tumors
Abstract
DNMT1, the most abundant human methyltransferase, is responsible for translating the correct methylation pattern during DNA replication, and aberrant methylation by DNMT1 has been linked to tumorigenesis. We have developed a sensitive signal-on electrochemical assay for the measurement of DNMT1 activity in crude tissue lysates. We have further analyzed ten tumor sets and have found a direct correlation between DNMT1 hyperactivity and tumorous tissue. In the majority of samples analyzed, the tumorous tissue has significantly higher DNMT1 activity than the healthy adjacent tissue. No such correlation is observed in measurements of DNMT1 expression by qPCR, DNMT1 protein abundance by western blotting, or DNMT1 activity using a radiometric DNA labeling assay. DNMT1 hyperactivity can result from both protein overexpression and enzyme hyperactivity. DNMT1 activity measured electrochemically provides a direct measure of activity in cell lysates and, as a result, provides a sensitive and early indication of cancerous transformation.
Copyright © 2015 Elsevier Ltd. All rights reserved.
Figures
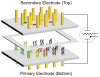



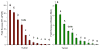

Comment in
-
DNMT1 and Cancer: An Electrifying Link.Chem Biol. 2015 Jul 23;22(7):810-1. doi: 10.1016/j.chembiol.2015.07.004. Chem Biol. 2015. PMID: 26207295
References
-
- Baylin SB, Herman JG. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends in Genetics. 2000;16:168–174. - PubMed
-
- Bestor TH. The DNA methyltransferases of mammals. Human Mol Genet. 2000;9:2395–2402. - PubMed
-
- De Marzo AM, Marchi VL, Yang ES, Veeraswamy R, Lin X, Nelson WG. Abnormal Regulation of DNA Methyltransferase Expression during Colorectal Carcinogenesis. Cancer Res. 1999;59:3855–3860. - PubMed
-
- Esteller M, Toyota M, Sanchez-Cespedes M, Capella G, Peinado MA, Watkins DN, Issa JPJ, Sidransky D, Baylin SB, Herman JG. Inactivation of the DNA Repair Gene O6-Methylguanine-DNA Methyltransferase by Promoter Hypermethylation Is Associated with G to A Mutations in K-ras in Colorectal Tumorigenesis. Cancer Res. 2000;60:2368–2371. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

