T-cell exhaustion, co-stimulation and clinical outcome in autoimmunity and infection
- PMID: 26123020
- PMCID: PMC4623162
- DOI: 10.1038/nature14468
T-cell exhaustion, co-stimulation and clinical outcome in autoimmunity and infection
Abstract
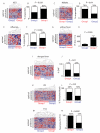
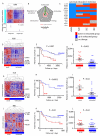
The clinical course of autoimmune and infectious disease varies greatly, even between individuals with the same condition. An understanding of the molecular basis for this heterogeneity could lead to significant improvements in both monitoring and treatment. During chronic infection the process of T-cell exhaustion inhibits the immune response, facilitating viral persistence. Here we show that a transcriptional signature reflecting CD8 T-cell exhaustion is associated with poor clearance of chronic viral infection, but conversely predicts better prognosis in multiple autoimmune diseases. The development of CD8 T-cell exhaustion during chronic infection is driven both by persistence of antigen and by a lack of accessory 'help' signals. In autoimmunity, we find that where evidence of CD4 T-cell co-stimulation is pronounced, that of CD8 T-cell exhaustion is reduced. We can reproduce the exhaustion signature by modifying the balance of persistent stimulation of T-cell antigen receptors and specific CD2-induced co-stimulation provided to human CD8 T cells in vitro, suggesting that each process plays a role in dictating outcome in autoimmune disease. The 'non-exhausted' T-cell state driven by CD2-induced co-stimulation is reduced by signals through the exhaustion-associated inhibitory receptor PD-1, suggesting that induction of exhaustion may be a therapeutic strategy in autoimmune and inflammatory disease. Using expression of optimal surrogate markers of co-stimulation/exhaustion signatures in independent data sets, we confirm an association with good clinical outcome or response to therapy in infection (hepatitis C virus) and vaccination (yellow fever, malaria, influenza), but poor outcome in autoimmune and inflammatory disease (type 1 diabetes, anti-neutrophil cytoplasmic antibody-associated vasculitis, systemic lupus erythematosus, idiopathic pulmonary fibrosis and dengue haemorrhagic fever). Thus, T-cell exhaustion plays a central role in determining outcome in autoimmune disease and targeted manipulation of this process could lead to new therapeutic opportunities.
Figures

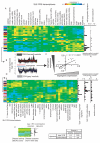
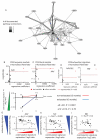
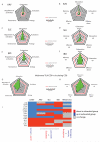
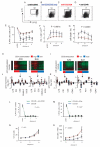







Comment in
-
Autoimmunity: Benefits of exhaustion.Nat Rev Immunol. 2015 Aug;15(8):468. doi: 10.1038/nri3890. Epub 2015 Jul 10. Nat Rev Immunol. 2015. PMID: 26160615 No abstract available.
-
Immunology: T-cell exhaustion limits immune reactivity and is associated with good prognosis in autoimmune disease.Nat Rev Nephrol. 2015 Sep;11(9):503. doi: 10.1038/nrneph.2015.115. Epub 2015 Jul 14. Nat Rev Nephrol. 2015. PMID: 26168738 No abstract available.
-
Immunology: T-cell exhaustion limits immune reactivity and is associated with good prognosis in autoimmune disease.Nat Rev Rheumatol. 2015 Sep;11(9):501. doi: 10.1038/nrrheum.2015.101. Epub 2015 Jul 14. Nat Rev Rheumatol. 2015. PMID: 26168912 No abstract available.
References
-
- Wherry EJ. T cell exhaustion. Nature immunology. 2011;12:492–499. - PubMed
METHODS REFERENCES
-
- Stone JH, et al. A disease-specific activity index for Wegener’s granulomatosis: modification of the Birmingham Vasculitis Activity Score. International Network for the Study of the Systemic Vasculitides (INSSYS) Arthritis and rheumatism. 2001;44:912–920. - PubMed
-
- Tan EM, et al. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis and rheumatism. 1982;25:1271–1277. - PubMed
-
- Isenberg DA, et al. BILAG 2004. Development and initial validation of an updated version of the British Isles Lupus Assessment Group’s disease activity index for patients with systemic lupus erythematosus. Rheumatology. 2005;44:902–906. - PubMed
-
- Silverberg MS, et al. Toward an integrated clinical, molecular and serological classification of inflammatory bowel disease: report of a Working Party of the 2005 Montreal World Congress of Gastroenterology. Can J Gastroenterol. 2005;19(Suppl A):5A–36A. - PubMed
-
- Harvey RF, Bradshaw MJ. Measuring Crohn’s disease activity. Lancet. 1980;1:1134–1135. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

