Circadian clock gene LATE ELONGATED HYPOCOTYL directly regulates the timing of floral scent emission in Petunia
- PMID: 26124104
- PMCID: PMC4534231
- DOI: 10.1073/pnas.1422875112
Circadian clock gene LATE ELONGATED HYPOCOTYL directly regulates the timing of floral scent emission in Petunia
Abstract
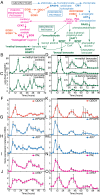
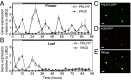
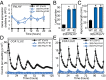
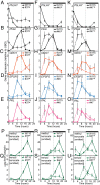
Flowers present a complex display of signals to attract pollinators, including the emission of floral volatiles. Volatile emission is highly regulated, and many species restrict emissions to specific times of the day. This rhythmic emission of scent is regulated by the circadian clock; however, the mechanisms have remained unknown. In Petunia hybrida, volatile emissions are dominated by products of the floral volatile benzenoid/phenylpropanoid (FVBP) metabolic pathway. Here we demonstrate that the circadian clock gene P. hybrida LATE ELONGATED HYPOCOTYL (LHY; PhLHY) regulates the daily expression patterns of the FVBP pathway genes and floral volatile production. PhLHY expression peaks in the morning, antiphasic to the expression of P. hybrida GIGANTEA (PhGI), the master scent regulator ODORANT1 (ODO1), and many other evening-expressed FVBP genes. Overexpression phenotypes of PhLHY in Arabidopsis caused an arrhythmic clock phenotype, which resembles those of LHY overexpressors. In Petunia, constitutive expression of PhLHY depressed the expression levels of PhGI, ODO1, evening-expressed FVBP pathway genes, and FVBP emission in flowers. Additionally, in the Petunia lines in which PhLHY expression was reduced, the timing of peak expression of PhGI, ODO1, and the FVBP pathway genes advanced to the morning. Moreover, PhLHY protein binds to cis-regulatory elements called evening elements that exist in promoters of ODO1 and other FVBP genes. Thus, our results imply that PhLHY directly sets the timing of floral volatile emission by restricting the expression of ODO1 and other FVBP genes to the evening in Petunia.
Keywords: LHY; Petunia hybrida; benzenoids; circadian rhythm; floral volatile.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Harmer SL. The circadian system in higher plants. Annu Rev Plant Biol. 2009;60:357–377. - PubMed
-
- Dodd AN, et al. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science. 2005;309(5734):630–633. - PubMed
-
- Muhlemann JK, Klempien A, Dudareva N. Floral volatiles: From biosynthesis to function. Plant Cell Environ. 2014;37(8):1936–1949. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

