Intrinsic unfoldase/foldase activity of the chaperonin GroEL directly demonstrated using multinuclear relaxation-based NMR
- PMID: 26124125
- PMCID: PMC4517251
- DOI: 10.1073/pnas.1510083112
Intrinsic unfoldase/foldase activity of the chaperonin GroEL directly demonstrated using multinuclear relaxation-based NMR
Abstract
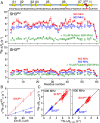
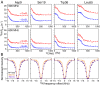
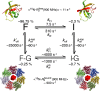
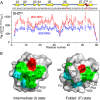
The prototypical chaperonin GroEL assists protein folding through an ATP-dependent encapsulation mechanism. The details of how GroEL folds proteins remain elusive, particularly because encapsulation is not an absolute requirement for successful re/folding. Here we make use of a metastable model protein substrate, comprising a triple mutant of Fyn SH3, to directly demonstrate, by simultaneous analysis of three complementary NMR-based relaxation experiments (lifetime line broadening, dark state exchange saturation transfer, and Carr-Purcell-Meinboom-Gill relaxation dispersion), that apo GroEL accelerates the overall interconversion rate between the native state and a well-defined folding intermediate by about 20-fold, under conditions where the "invisible" GroEL-bound states have occupancies below 1%. This is largely achieved through a 500-fold acceleration in the folded-to-intermediate transition of the protein substrate. Catalysis is modulated by a kinetic deuterium isotope effect that reduces the overall interconversion rate between the GroEL-bound species by about 3-fold, indicative of a significant hydrophobic contribution. The location of the GroEL binding site on the folding intermediate, mapped from (15)N, (1)HN, and (13)Cmethyl relaxation dispersion experiments, is composed of a prominent, surface-exposed hydrophobic patch.
Keywords: chaperonins; dark state exchange saturation transfer; invisible states; lifetime line broadening; relaxation dispersion.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

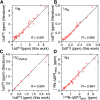

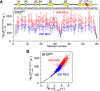
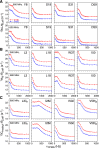

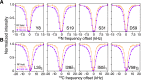
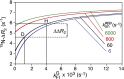
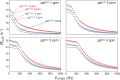
Comment in
-
Reply to Marchenko et al.: Flux analysis of GroEL-assisted protein folding/unfolding.Proc Natl Acad Sci U S A. 2015 Dec 15;112(50):E6833-4. doi: 10.1073/pnas.1520474112. Epub 2015 Nov 24. Proc Natl Acad Sci U S A. 2015. PMID: 26604310 Free PMC article. No abstract available.
-
Strict experimental evidence that apo-chaperonin GroEL does not accelerate protein folding, although it does accelerate one of its steps.Proc Natl Acad Sci U S A. 2015 Dec 15;112(50):E6831-2. doi: 10.1073/pnas.1517712112. Epub 2015 Nov 24. Proc Natl Acad Sci U S A. 2015. PMID: 26604318 Free PMC article. No abstract available.
References
-
- Dobson CM. Protein folding and misfolding. Nature. 2003;426(6968):884–890. - PubMed
-
- Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475(7356):324–332. - PubMed
-
- Thirumalai D, Lorimer GH. Chaperonin-mediated protein folding. Ann Rev Biophys Biomol Struct. 2001;30:245–269. - PubMed
-
- Saibil HR. Chaperone machines in action. Curr Opin Struct Biol. 2008;18(1):35–42. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

