Sex hormone-dependent tRNA halves enhance cell proliferation in breast and prostate cancers
- PMID: 26124144
- PMCID: PMC4517238
- DOI: 10.1073/pnas.1510077112
Sex hormone-dependent tRNA halves enhance cell proliferation in breast and prostate cancers
Abstract
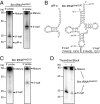
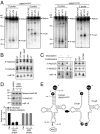
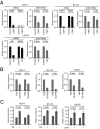
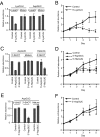
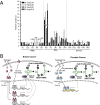
Sex hormones and their receptors play critical roles in the development and progression of the breast and prostate cancers. Here we report that a novel type of transfer RNA (tRNA)-derived small RNA, termed Sex HOrmone-dependent TRNA-derived RNAs (SHOT-RNAs), are specifically and abundantly expressed in estrogen receptor (ER)-positive breast cancer and androgen receptor (AR)-positive prostate cancer cell lines. SHOT-RNAs are not abundantly present in ER(-) breast cancer, AR(-) prostate cancer, or other examined cancer cell lines from other tissues. ER-dependent accumulation of SHOT-RNAs is not limited to a cell culture system, but it also occurs in luminal-type breast cancer patient tissues. SHOT-RNAs are produced from aminoacylated mature tRNAs by angiogenin-mediated anticodon cleavage, which is promoted by sex hormones and their receptors. Resultant 5'- and 3'-SHOT-RNAs, corresponding to 5'- and 3'-tRNA halves, bear a cyclic phosphate (cP) and an amino acid at the 3'-end, respectively. By devising a "cP-RNA-seq" method that is able to exclusively amplify and sequence cP-containing RNAs, we identified the complete repertoire of 5'-SHOT-RNAs. Furthermore, 5'-SHOT-RNA, but not 3'-SHOT-RNA, has significant functional involvement in cell proliferation. These results have unveiled a novel tRNA-engaged pathway in tumorigenesis of hormone-dependent cancers and implicate SHOT-RNAs as potential candidates for biomarkers and therapeutic targets.
Keywords: breast cancer; prostate cancer; sex hormone; tRNA; tRNA half.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Comment in
-
Re: Sex Hormone-Dependent tRNA Halves Enhance Cell Proliferation in Breast and Prostate Cancers.J Urol. 2016 Apr;195(4 Pt 1):1168-9. doi: 10.1016/j.juro.2016.01.019. Epub 2016 Jan 21. J Urol. 2016. PMID: 27302822 No abstract available.
-
Re: Mutations in TERT Promoter and FGFR3 and Telomere Length in Bladder Cancer.J Urol. 2016 Apr;195(4 Pt 1):1168-9. doi: 10.1016/j.juro.2016.01.020. Epub 2016 Jan 21. J Urol. 2016. PMID: 27302823 No abstract available.
References
-
- Gingold H, et al. A dual program for translation regulation in cellular proliferation and differentiation. Cell. 2014;158(6):1281–1292. - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

