RNA-Seq analysis of isolate- and growth phase-specific differences in the global transcriptomes of enteropathogenic Escherichia coli prototype isolates
- PMID: 26124752
- PMCID: PMC4464170
- DOI: 10.3389/fmicb.2015.00569
RNA-Seq analysis of isolate- and growth phase-specific differences in the global transcriptomes of enteropathogenic Escherichia coli prototype isolates
Abstract
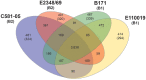
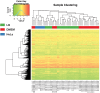
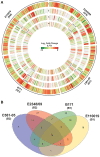
Enteropathogenic Escherichia coli (EPEC) are a leading cause of diarrheal illness among infants in developing countries. E. coli isolates classified as typical EPEC are identified by the presence of the locus of enterocyte effacement (LEE) and the bundle-forming pilus (BFP), and absence of the Shiga-toxin genes, while the atypical EPEC also encode LEE but do not encode BFP or Shiga-toxin. Comparative genomic analyses have demonstrated that EPEC isolates belong to diverse evolutionary lineages and possess lineage- and isolate-specific genomic content. To investigate whether this genomic diversity results in significant differences in global gene expression, we used an RNA sequencing (RNA-Seq) approach to characterize the global transcriptomes of the prototype typical EPEC isolates E2348/69, B171, C581-05, and the prototype atypical EPEC isolate E110019. The global transcriptomes were characterized during laboratory growth in two different media and three different growth phases, as well as during adherence of the EPEC isolates to human cells using in vitro tissue culture assays. Comparison of the global transcriptomes during these conditions was used to identify isolate- and growth phase-specific differences in EPEC gene expression. These analyses resulted in the identification of genes that encode proteins involved in survival and metabolism that were coordinately expressed with virulence factors. These findings demonstrate there are isolate- and growth phase-specific differences in the global transcriptomes of EPEC prototype isolates, and highlight the utility of comparative transcriptomics for identifying additional factors that are directly or indirectly involved in EPEC pathogenesis.
Keywords: RNA-sequencing; diversity; enteropathogenic Escherichia coli; pathogenesis; regulation.
Figures



References
-
- Abu-Ali G. S., Ouellette L. M., Henderson S. T., Lacher D. W., Riordan J. T., Whittam T. S., et al. (2010). Increased adherence and expression of virulence genes in a lineage of Escherichia coli O157:H7 commonly associated with human infections. PLoS ONE 5:e10167. 10.1371/journal.pone.0010167 - DOI - PMC - PubMed
-
- Bhatt S., Edwards A. N., Nguyen H. T., Merlin D., Romeo T., Kalman D. (2009). The RNA binding protein CsrA is a pleiotropic regulator of the locus of enterocyte effacement pathogenicity island of enteropathogenic Escherichia coli. Infect. Immun. 77, 3552–3568. 10.1128/IAI.00418-09 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

