Circadian rhythms and post-transcriptional regulation in higher plants
- PMID: 26124767
- PMCID: PMC4464108
- DOI: 10.3389/fpls.2015.00437
Circadian rhythms and post-transcriptional regulation in higher plants
Abstract
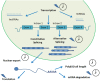
The circadian clock of plants allows them to cope with daily changes in their environment. This is accomplished by the rhythmic regulation of gene expression, in a process that involves many regulatory steps. One of the key steps involved at the RNA level is post-transcriptional regulation, which ensures a correct control on the different amounts and types of mRNA that will ultimately define the current physiological state of the plant cell. Recent advances in the study of the processes of regulation of pre-mRNA processing, RNA turn-over and surveillance, regulation of translation, function of lncRNAs, biogenesis and function of small RNAs, and the development of bioinformatics tools have helped to vastly expand our understanding of how this regulatory step performs its role. In this work we review the current progress in circadian regulation at the post-transcriptional level research in plants. It is the continuous interaction of all the information flow control post-transcriptional processes that allow a plant to precisely time and predict daily environmental changes.
Keywords: Arabidopsis thaliana; RNA turnover; alternative splicing; circadian rhythms; mRNA nuclear export; polyadenilation; post-transcriptional regulation; regulation of translation.
Figures


References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

