Analysis of the type II-A CRISPR-Cas system of Streptococcus agalactiae reveals distinctive features according to genetic lineages
- PMID: 26124774
- PMCID: PMC4466440
- DOI: 10.3389/fgene.2015.00214
Analysis of the type II-A CRISPR-Cas system of Streptococcus agalactiae reveals distinctive features according to genetic lineages
Abstract
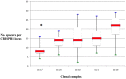
CRISPR-Cas systems (clustered regularly interspaced short palindromic repeats/CRISPR-associated proteins) are found in 90% of archaea and about 40% of bacteria. In this original system, CRISPR arrays comprise short, almost unique sequences called spacers that are interspersed with conserved palindromic repeats. These systems play a role in adaptive immunity and participate to fight non-self DNA such as integrative and conjugative elements, plasmids, and phages. In Streptococcus agalactiae, a bacterium implicated in colonization and infections in humans since the 1960s, two CRISPR-Cas systems have been described. A type II-A system, characterized by proteins Cas9, Cas1, Cas2, and Csn2, is ubiquitous, and a type I-C system, with the Cas8c signature protein, is present in about 20% of the isolates. Unlike type I-C, which appears to be non-functional, type II-A appears fully functional. Here we studied type II-A CRISPR-cas loci from 126 human isolates of S. agalactiae belonging to different clonal complexes that represent the diversity of the species and that have been implicated in colonization or infection. The CRISPR-cas locus was analyzed both at spacer and repeat levels. Major distinctive features were identified according to the phylogenetic lineages previously defined by multilocus sequence typing, especially for the sequence type (ST) 17, which is considered hypervirulent. Among other idiosyncrasies, ST-17 shows a significantly lower number of spacers in comparison with other lineages. This characteristic could reflect the peculiar virulence or colonization specificities of this lineage.
Keywords: CRISPR-Cas; ST-17; Streptococcus agalactiae; phylogeny; typing.
Figures


References
-
- Bohnsack J. F., Whiting A., Gottschalk M., Dunn D. M., Weiss R., Azimi P. H., et al. (2008). Population structure of invasive and colonizing strains of Streptococcus agalactiae from neonates of six U.S. Academic Centers from 1995 to 1999. J. Clin. Microbiol. 46 1285–1291. 10.1128/JCM.02105-07 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

