Use of RNA sequencing to evaluate rheumatic disease patients
- PMID: 26126608
- PMCID: PMC4488125
- DOI: 10.1186/s13075-015-0677-3
Use of RNA sequencing to evaluate rheumatic disease patients
Abstract
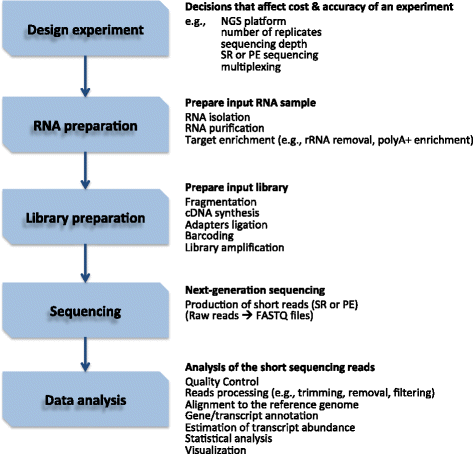
Studying the factors that control gene expression is of substantial importance for rheumatic diseases with poorly understood etiopathogenesis. In the past, gene expression microarrays have been used to measure transcript abundance on a genome-wide scale in a particular cell, tissue or organ. Microarray analysis has led to gene signatures that differentiate rheumatic diseases, and stages of a disease, as well as response to treatments. Nowadays, however, with the advent of next-generation sequencing methods, massive parallel sequencing of RNA tends to be the technology of choice for gene expression profiling, due to several advantages over microarrays, as well as for the detection of non-coding transcripts and alternative splicing events. In this review, we describe how RNA sequencing enables unbiased interrogation of the abundance and complexity of the transcriptome, and present a typical experimental workflow and bioinformatics tools that are often used for RNA sequencing analysis. We also discuss different uses of this next-generation sequencing technology to evaluate rheumatic disease patients and investigate the pathogenesis of rheumatic diseases such as rheumatoid arthritis, systemic lupus erythematosus, juvenile idiopathic arthritis and Sjögren's syndrome.
Figures
References
-
- van der Pouw Kraan TC, van Gaalen FA, Huizinga TW, Pieterman E, Breedveld FC, Verweij CL. Discovery of distinctive gene expression profiles in rheumatoid synovium using cDNA microarray technology: evidence for the existence of multiple pathways of tissue destruction and repair. Genes Immun. 2003;4:187–96. doi: 10.1038/sj.gene.6363975. - DOI - PubMed
-
- van der Pouw Kraan TC, van Gaalen FA, Kasperkovitz PV, Verbeet NL, Smeets TJ, Kraan MC, et al. Rheumatoid arthritis is a heterogeneous disease: evidence for differences in the activation of the STAT-1 pathway between rheumatoid tissues. Arthritis Rheum. 2003;48:2132–45. doi: 10.1002/art.11096. - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

