Core protein: A pleiotropic keystone in the HBV lifecycle
- PMID: 26129969
- PMCID: PMC4537649
- DOI: 10.1016/j.antiviral.2015.06.020
Core protein: A pleiotropic keystone in the HBV lifecycle
Abstract
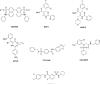
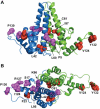
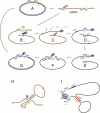
Hepatitis B Virus (HBV) is a small virus whose genome has only four open reading frames. We argue that the simplicity of the virion correlates with a complexity of functions for viral proteins. We focus on the HBV core protein (Cp), a small (183 residue) protein that self-assembles to form the viral capsid. However, its functions are a little more complicated than that. In an infected cell Cp modulates almost every step of the viral lifecycle. Cp is bound to nuclear viral DNA and affects its epigenetics. Cp correlates with RNA specificity. Cp assembles specifically on a reverse transcriptase-viral RNA complex or, apparently, nothing at all. Indeed Cp has been one of the model systems for investigation of virus self-assembly. Cp participates in regulation of reverse transcription. Cp signals completion of reverse transcription to support virus secretion. Cp carries both nuclear localization signals and HBV surface antigen (HBsAg) binding sites; both of these functions appear to be regulated by contents of the capsid. Cp can be targeted by antivirals - while self-assembly is the most accessible of Cp activities, we argue that it makes sense to engage the broader spectrum of Cp function. This article forms part of a symposium in Antiviral Research on "From the discovery of the Australia antigen to the development of new curative therapies for hepatitis B: an unfinished story."
Keywords: Antiviral therapy; Capsid; Hepatitis B Virus; Self-assembly.
Copyright © 2015 Elsevier B.V. All rights reserved.
Figures
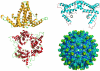

References
-
- EASL clinical practice guidelines: Management of chronic hepatitis B virus infection. J Hepatol. 2012;57:167–185. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

