Biomarker discovery: quantification of microRNAs and other small non-coding RNAs using next generation sequencing
- PMID: 26130076
- PMCID: PMC4487992
- DOI: 10.1186/s12920-015-0109-x
Biomarker discovery: quantification of microRNAs and other small non-coding RNAs using next generation sequencing
Abstract
Background: Small ncRNAs (sncRNAs) offer great hope as biomarkers of disease and response to treatment. This has been highlighted in the context of several medical conditions such as cancer, liver disease, cardiovascular disease, and central nervous system disorders, among many others. Here we assessed several steps involved in the development of an ncRNA biomarker discovery pipeline, ranging from sample preparation to bioinformatic processing of small RNA sequencing data.
Methods: A total of 45 biological samples were included in the present study. All libraries were prepared using the Illumina TruSeq Small RNA protocol and sequenced using the HiSeq2500 or MiSeq Illumina sequencers. Small RNA sequencing data was validated using qRT-PCR. At each stage, we evaluated the pros and cons of different techniques that may be suitable for different experimental designs. Evaluation methods included quality of data output in relation to hands-on laboratory time, cost, and efficiency of processing.
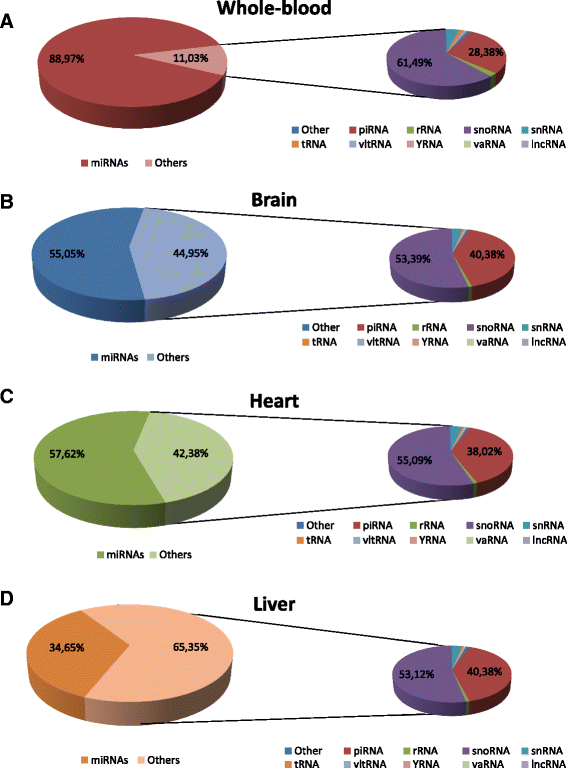
Results: Our results show that good quality sequencing libraries can be prepared from small amounts of total RNA and that varying degradation levels in the samples do not have a significant effect on the overall quantification of sncRNAs via NGS. In addition, we describe the strengths and limitations of three commercially available library preparation methods: (1) Novex TBE PAGE gel; (2) Pippin Prep automated gel system; and (3) AMPure XP beads. We describe our bioinformatics pipeline, provide recommendations for sequencing coverage, and describe in detail the expression and distribution of all sncRNAs in four human tissues: whole-blood, brain, heart and liver.
Conclusions: Ultimately this study provides tools and outcome metrics that will aid researchers and clinicians in choosing an appropriate and effective high-throughput sequencing quantification method for various study designs, and overall generating valuable information that can contribute to our understanding of small ncRNAs as potential biomarkers and mediators of biological functions and disease.
Figures






References
-
- Davis J, Maes M, Andreazza A, McGrath JJ, Tye SJ, Berk M. Towards a classification of biomarkers of neuropsychiatric disease: from encompass to compass. Mol Psychiatry. 2014 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

