Two separable functions of Ctp1 in the early steps of meiotic DNA double-strand break repair
- PMID: 26130711
- PMCID: PMC4551917
- DOI: 10.1093/nar/gkv644
Two separable functions of Ctp1 in the early steps of meiotic DNA double-strand break repair
Abstract
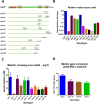
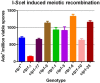
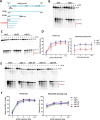
Meiotic programmed DNA double-strand break (DSB) repair is essential for crossing-over and viable gamete formation and requires removal of Spo11-oligonucleotide complexes from 5' ends (clipping) and their resection to generate invasive 3'-end single-stranded DNA (resection). Ctp1 (Com1, Sae2, CtIP homolog) acting with the Mre11-Rad50-Nbs1 (MRN) complex is required in both steps. We isolated multiple S. pombe ctp1 mutants deficient in clipping but proficient in resection during meiosis. Remarkably, all of the mutations clustered in or near the conserved CxxC or RHR motif in the C-terminal portion. The mutants tested, like ctp1Δ, were clipping-deficient by both genetic and physical assays-. But, unlike ctp1Δ, these mutants were recombination-proficient for Rec12 (Spo11 homolog)-independent break-repair and resection-proficient by physical assay. We conclude that the intracellular Ctp1 C-terminal portion is essential for clipping, while the N-terminal portion is sufficient for DSB end-resection. This conclusion agrees with purified human CtIP resection and endonuclease activities being independent. Our mutants provide intracellular evidence for separable functions of Ctp1. Some mutations truncate Ctp1 in the same region as one of the CtIP mutations linked to the Seckel and Jawad severe developmental syndromes, suggesting that these syndromes are caused by a lack of clipping at DSB ends that require repair.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
-
- Keeney S. In: Recombination and meiosis: crossing-over and disjunction. Egel R, Lankenau D.-H., editors. Berlin: Springer-Verlag; 2007. pp. 81–123.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

