Human DNA ligase III bridges two DNA ends to promote specific intermolecular DNA end joining
- PMID: 26130724
- PMCID: PMC4538836
- DOI: 10.1093/nar/gkv652
Human DNA ligase III bridges two DNA ends to promote specific intermolecular DNA end joining
Abstract
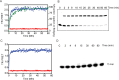
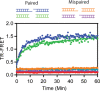
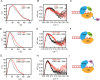
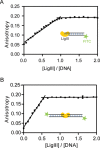
Mammalian DNA ligase III (LigIII) functions in both nuclear and mitochondrial DNA metabolism. In the nucleus, LigIII has functional redundancy with DNA ligase I whereas LigIII is the only mitochondrial DNA ligase and is essential for the survival of cells dependent upon oxidative respiration. The unique LigIII zinc finger (ZnF) domain is not required for catalytic activity but senses DNA strand breaks and stimulates intermolecular ligation of two DNAs by an unknown mechanism. Consistent with this activity, LigIII acts in an alternative pathway of DNA double strand break repair that buttresses canonical non-homologous end joining (NHEJ) and is manifest in NHEJ-defective cancer cells, but how LigIII acts in joining intermolecular DNA ends versus nick ligation is unclear. To investigate how LigIII efficiently joins two DNAs, we developed a real-time, fluorescence-based assay of DNA bridging suitable for high-throughput screening. On a nicked duplex DNA substrate, the results reveal binding competition between the ZnF and the oligonucleotide/oligosaccharide-binding domain, one of three domains constituting the LigIII catalytic core. In contrast, these domains collaborate and are essential for formation of a DNA-bridging intermediate by adenylated LigIII that positions a pair of blunt-ended duplex DNAs for efficient and specific intermolecular ligation.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




References
-
- Tomkinson A.E., Vijayakumar S., Pascal J.M., Ellenberger T. DNA ligases: structure, reaction mechanism, and function. Chem. Rev. 2006;106:687–699. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

