CRISPR-Cas9 Genome Editing in Drosophila
- PMID: 26131852
- PMCID: PMC4506758
- DOI: 10.1002/0471142727.mb3102s111
CRISPR-Cas9 Genome Editing in Drosophila
Abstract
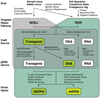
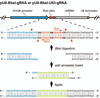
The CRISPR-Cas9 system has transformed genome engineering of model organisms from possible to practical. CRISPR-Cas9 can be readily programmed to generate sequence-specific double-strand breaks that disrupt targeted loci when repaired by error-prone non-homologous end joining (NHEJ) or to catalyze precise genome modification through homology-directed repair (HDR). Here we describe a streamlined approach for rapid and highly efficient engineering of the Drosophila genome via CRISPR-Cas9-mediated HDR. In this approach, transgenic flies expressing Cas9 are injected with plasmids to express guide RNAs (gRNAs) and positively marked donor templates. We detail target-site selection; gRNA plasmid generation; donor template design and construction; and the generation, identification, and molecular confirmation of engineered lines. We also present alternative approaches and highlight key considerations for experimental design. The approach outlined here can be used to rapidly and reliably generate a variety of engineered modifications, including genomic deletions and replacements, precise sequence edits, and incorporation of protein tags.
Keywords: CRISPR; Cas9; Drosophila; genome engineering; homology directed repair.
Copyright © 2015 John Wiley & Sons, Inc.
Figures


References
-
- Bassett AR, Liu JL. CRISPR/Cas9 and genome editing in Drosophila. Journal of genetics and genomics = Yi chuan xue bao. 2014;41:7–19. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

