Lenalidomide induces ubiquitination and degradation of CK1α in del(5q) MDS
- PMID: 26131937
- PMCID: PMC4853910
- DOI: 10.1038/nature14610
Lenalidomide induces ubiquitination and degradation of CK1α in del(5q) MDS
Abstract
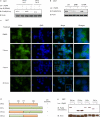
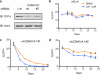
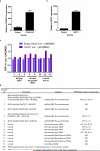
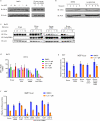
Lenalidomide is a highly effective treatment for myelodysplastic syndrome (MDS) with deletion of chromosome 5q (del(5q)). Here, we demonstrate that lenalidomide induces the ubiquitination of casein kinase 1A1 (CK1α) by the E3 ubiquitin ligase CUL4-RBX1-DDB1-CRBN (known as CRL4(CRBN)), resulting in CK1α degradation. CK1α is encoded by a gene within the common deleted region for del(5q) MDS and haploinsufficient expression sensitizes cells to lenalidomide therapy, providing a mechanistic basis for the therapeutic window of lenalidomide in del(5q) MDS. We found that mouse cells are resistant to lenalidomide but that changing a single amino acid in mouse Crbn to the corresponding human residue enables lenalidomide-dependent degradation of CK1α. We further demonstrate that minor side chain modifications in thalidomide and a novel analogue, CC-122, can modulate the spectrum of substrates targeted by CRL4(CRBN). These findings have implications for the clinical activity of lenalidomide and related compounds, and demonstrate the therapeutic potential of novel modulators of E3 ubiquitin ligases.
Figures






Comment in
-
Myeloid disease: Another action of a thalidomide derivative.Nature. 2015 Jul 9;523(7559):167-8. doi: 10.1038/nature14628. Epub 2015 Jul 1. Nature. 2015. PMID: 26131932 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

