mTOR Directs Breast Morphogenesis through the PKC-alpha-Rac1 Signaling Axis
- PMID: 26132202
- PMCID: PMC4488502
- DOI: 10.1371/journal.pgen.1005291
mTOR Directs Breast Morphogenesis through the PKC-alpha-Rac1 Signaling Axis
Abstract
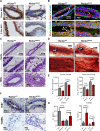
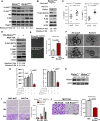
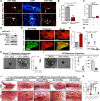
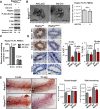
Akt phosphorylation is a major driver of cell survival, motility, and proliferation in development and disease, causing increased interest in upstream regulators of Akt like mTOR complex 2 (mTORC2). We used genetic disruption of Rictor to impair mTORC2 activity in mouse mammary epithelia, which decreased Akt phosphorylation, ductal length, secondary branching, cell motility, and cell survival. These effects were recapitulated with a pharmacological dual inhibitor of mTORC1/mTORC2, but not upon genetic disruption of mTORC1 function via Raptor deletion. Surprisingly, Akt re-activation was not sufficient to rescue cell survival or invasion, and modestly increased branching of mTORC2-impaired mammary epithelial cells (MECs) in culture and in vivo. However, another mTORC2 substrate, protein kinase C (PKC)-alpha, fully rescued mTORC2-impaired MEC branching, invasion, and survival, as well as branching morphogenesis in vivo. PKC-alpha-mediated signaling through the small GTPase Rac1 was necessary for mTORC2-dependent mammary epithelial development during puberty, revealing a novel role for Rictor/mTORC2 in MEC survival and motility during branching morphogenesis through a PKC-alpha/Rac1-dependent mechanism.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Sarbassov D.D., et al., Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell, 2006. 22(2): p. 159–68. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

