Properties of targeted preamplification in DNA and cDNA quantification
- PMID: 26132215
- PMCID: PMC4673511
- DOI: 10.1586/14737159.2015.1057124
Properties of targeted preamplification in DNA and cDNA quantification
Abstract
Objective: Quantification of small molecule numbers often requires preamplification to generate enough copies for accurate downstream enumerations. Here, we studied experimental parameters in targeted preamplification and their effects on downstream quantitative real-time PCR (qPCR).
Methods: To evaluate different strategies, we monitored the preamplification reaction in real-time using SYBR Green detection chemistry followed by melting curve analysis. Furthermore, individual targets were evaluated by qPCR.
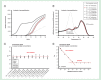
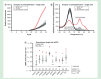
Result: The preamplification reaction performed best when a large number of primer pairs was included in the primer pool. In addition, preamplification efficiency, reproducibility and specificity were found to depend on the number of template molecules present, primer concentration, annealing time and annealing temperature. The amount of nonspecific PCR products could also be reduced about 1000-fold using bovine serum albumin, glycerol and formamide in the preamplification.
Conclusion: On the basis of our findings, we provide recommendations how to perform robust and highly accurate targeted preamplification in combination with qPCR or next-generation sequencing.
Keywords: experimental design; multiplex PCR; preamplification; primer-pools; quantitative real-time PCR; single-cell analysis; targeted preamplification.
Figures






References
-
- Guo G, Huss M, Tong GQ, et al. Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev Cell. 2010;18(4):675–85. - PubMed
-
- Norrman K, Strombeck A, Semb H, Stahlberg A. Distinct gene expression signatures in human embryonic stem cells differentiated towards definitive endoderm at single-cell level. Methods. 2012;59(1):59–70. - PubMed
-
- Bengtsson M, Stahlberg A, Rorsman P, Kubista M. Gene expression profiling in single cells from the pancreatic islets of Langerhans reveals lognormal distribution of mRNA levels. Genome Res. 2005;15(10):1388–92. - PMC - PubMed
-
Single-cell study revealing that transcript levels have lognormal expression features in mammalian cells.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
