Differences between Cryptococcus neoformans and Cryptococcus gattii in the Molecular Mechanisms Governing Utilization of D-Amino Acids as the Sole Nitrogen Source
- PMID: 26132227
- PMCID: PMC4489021
- DOI: 10.1371/journal.pone.0131865
Differences between Cryptococcus neoformans and Cryptococcus gattii in the Molecular Mechanisms Governing Utilization of D-Amino Acids as the Sole Nitrogen Source
Abstract
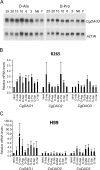
The ability to grow on media containing certain D-amino acids as a sole nitrogen source is widely utilized to differentiate Cryptococcus gattii from C. neoformans. We used the C. neoformans H99 and C. gattii R265 strains to dissect the mechanisms of D-amino acids utilization. We identified three putative D-amino acid oxidase (DAO) genes in both strains and showed that each DAO gene plays different roles in D-amino acid utilization in each strain. Deletion of DAO2 retarded growth of R265 on eleven D-amino acids suggesting its prominent role on D-amino acid assimilation in R265. All three R265 DAO genes contributed to growth on D-Asn and D-Asp. DAO3 was required for growth and detoxification of D-Glu by both R265 and H99. Although growth of H99 on most D-amino acids was poor, deletion of DAO1 or DAO3 further exacerbated it on four D-amino acids. Overexpression of DAO2 or DAO3 enabled H99 to grow robustly on several D-amino acids suggesting that expression levels of the native DAO genes in H99 were insufficient for growth on D-amino acids. Replacing the H99 DAO2 gene with a single copy of the R265 DAO2 gene also enabled its utilization of several D-amino acids. Results of gene and promoter swaps of the DAO2 genes suggested that enzymatic activity of Dao2 in H99 might be lower compared to the R265 strain. A reduction in virulence was only observed when all DAO genes were deleted in R265 but not in H99 indicating a pathobiologically exclusive role of the DAO genes in R265. These results suggest that C. neoformans and C. gattii divergently evolved in D-amino acid utilization influenced by their major ecological niches.
Conflict of interest statement
Figures





References
-
- Heitman J, Kozel TR, Kwon-Chung J, Perfect JR, Casadevall A, editors (2011) Cryptococcus: from human pathogen to model yeast. Washington DC: ASM press; 620 p.
-
- Kavanaugh LA, Fraser JA, Dietrich FS (2006) Recent evolution of the human pathogen Cryptococcus neoformans by intervarietal transfer of a 14-gene fragment. Mol Biol Evol 23: 1879–1890. - PubMed
-
- Kwon-Chung KJ, Bennett JE (1992) Medical Mycology. Philadelphia: Lea & Febiger.
-
- Casadevall A, Perfect JR (1998) Cryptococcus neoformans. Washington, DC: ASM press.
-
- Fraser JA, Giles SS, Wenink EC, Geunes-Boyer SG, Wright JR, Diezmann S, et al. (2005) Same-sex mating and the origin of the Vancouver Island Cryptococcus gattii outbreak. Nature 437: 1360–1364. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

