Metabarcoding Analysis of Fungal Diversity in the Phyllosphere and Carposphere of Olive (Olea europaea)
- PMID: 26132745
- PMCID: PMC4489200
- DOI: 10.1371/journal.pone.0131069
Metabarcoding Analysis of Fungal Diversity in the Phyllosphere and Carposphere of Olive (Olea europaea)
Abstract
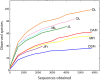
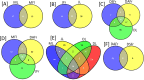
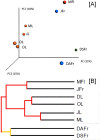
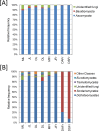
The fungal diversity associated with leaves, flowers and fruits of olive (Olea europaea) was investigated in different phenological stages (May, June, October and December) using an implemented metabarcoding approach. It consisted of the 454 pyrosequencing of the fungal ITS2 region and the subsequent phylogenetic analysis of relevant genera along with validated reference sequences. Most sequences were identified up to the species level or were associated with a restricted number of related taxa enabling supported speculations regarding their biological role. Analyses revealed a rich fungal community with 195 different OTUs. Ascomycota was the dominating phyla representing 93.6% of the total number of detected sequences followed by unidentified fungi (3.6%) and Basidiomycota (2.8%). A higher level of diversity was revealed for leaves compared to flowers and fruits. Among plant pathogens the genus Colletotrichum represented by three species (C. godetiae syn. C. clavatum, C. acutatum s.s and C. karstii) was the most abundant on ripe fruits but it was also detected in other organs. Pseudocercospora cladosporioides was detected with a high frequency in all leaf samples and to a less extent in ripe fruits. A much lower relative frequency was revealed for Spilocaea oleagina and for other putative pathogens including Fusarium spp., Neofusicoccum spp., and Alternaria spp. Among non-pathogen taxa, Aureobasidium pullulans, the species complex of Cladosporium cladosporioides and Devriesia spp. were the most represented. This study highlights the existence of a complex fungal consortium including both phytopathogenic and potentially antagonistic microorganisms that can have a significant impact on olive productions.
Conflict of interest statement
Figures


References
-
- Schena L, Agosteo GE, Cacciola SO. Olive diseases and disorders Kerala, India Transworld Research Network; 2011. 433 p.
-
- Agosteo GE, Schena L. Olive leaf spot In: Schena L, Agosteo GE, Cacciola SO, editors. Olive diseases and Disorders. 1 India Transworld Research Network; 2011. p. 143–76.
-
- Frisullo S, Carlucci A. Minor fungal diseases of olive In: Schena L, Agosteo GE, Cacciola SO, editors. Olive diseases and Disorders. India Transworld Research Network; 2011. p. 291–304.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

