Detection of skewed X-chromosome inactivation in Fragile X syndrome and X chromosome aneuploidy using quantitative melt analysis
- PMID: 26132880
- PMCID: PMC4836209
- DOI: 10.1017/erm.2015.11
Detection of skewed X-chromosome inactivation in Fragile X syndrome and X chromosome aneuploidy using quantitative melt analysis
Abstract
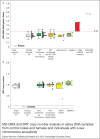
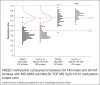
Methylation of the fragile X mental retardation 1 (FMR1) exon 1/intron 1 boundary positioned fragile X related epigenetic element 2 (FREE2), reveals skewed X-chromosome inactivation (XCI) in fragile X syndrome full mutation (FM: CGG > 200) females. XCI skewing has been also linked to abnormal X-linked gene expression with the broader clinical impact for sex chromosome aneuploidies (SCAs). In this study, 10 FREE2 CpG sites were targeted using methylation specific quantitative melt analysis (MS-QMA), including 3 sites that could not be analysed with previously used EpiTYPER system. The method was applied for detection of skewed XCI in FM females and in different types of SCA. We tested venous blood and saliva DNA collected from 107 controls (CGG < 40), and 148 FM and 90 SCA individuals. MS-QMA identified: (i) most SCAs if combined with a Y chromosome test; (ii) locus-specific XCI skewing towards the hypomethylated state in FM females; and (iii) skewed XCI towards the hypermethylated state in SCA with 3 or more X chromosomes, and in 5% of the 47,XXY individuals. MS-QMA output also showed significant correlation with the EpiTYPER reference method in FM males and females (P < 0.0001) and SCAs (P < 0.05). In conclusion, we demonstrate use of MS-QMA to quantify skewed XCI in two applications with diagnostic utility.
Figures

References
-
- Tost J, Gut I.G. (2012) DNA methylation analysis by MALDI mass spectrometry In Meyers: Encyclopedia of Molecular Cell Biology and Molecular Medicine: Epigenetic Regulation and Epigenomics (2nd edn) (Meyers R.A. ed.), pp. 1-40, Wiley-VCH Verlag GmbH & Co. KGaA.
-
- Godler D.E. et al. (2012) Fragile X mental retardation 1 (FMR1) intron 1 methylation in blood predicts verbal cognitive impairment in female carriers of expanded FMR1 alleles: evidence from a pilot study. Clinical Chemistry 58, 590-598 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

