Cryo-EM reveals the conformation of a substrate analogue in the human 20S proteasome core
- PMID: 26133119
- PMCID: PMC4506541
- DOI: 10.1038/ncomms8573
Cryo-EM reveals the conformation of a substrate analogue in the human 20S proteasome core
Abstract
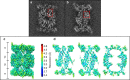
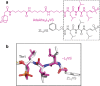
The proteasome is a highly regulated protease complex fundamental for cell homeostasis and controlled cell cycle progression. It functions by removing a wide range of specifically tagged proteins, including key cellular regulators. Here we present the structure of the human 20S proteasome core bound to a substrate analogue inhibitor molecule, determined by electron cryo-microscopy (cryo-EM) and single-particle analysis at a resolution of around 3.5 Å. Our map allows the building of protein coordinates as well as defining the location and conformation of the inhibitor at the different active sites. These results open new prospects to tackle the proteasome functional mechanisms. Moreover, they also further demonstrate that cryo-EM is emerging as a realistic approach for general structural studies of protein-ligand interactions.
Figures


References
-
- Lowe J. et al. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4A resolution. Science 268, 533–539 (1995). - PubMed
-
- Groll M. et al. Structure of 20S proteasome from yeast at 2.4A resolution. Nature 386, 463–471 (1997). - PubMed
-
- Unno M. et al. The structure of the mammalian 20S proteasome at 2.75A resolution. Structure 10, 609–618 (2002). - PubMed
-
- Huber E. M. et al. Immuno- and constitutive proteasome crystal structures reveal differences in substrate and inhibitor specificity. Cell 148, 727–738 (2012). - PubMed
-
- Groll M. et al. A gated channel into the proteasome core particle. Nat. Struct. Biol. 7, 1062–1067 (2000). - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

