Potential of promotion of alleles by genome editing to improve quantitative traits in livestock breeding programs
- PMID: 26133579
- PMCID: PMC4487592
- DOI: 10.1186/s12711-015-0135-3
Potential of promotion of alleles by genome editing to improve quantitative traits in livestock breeding programs
Erratum in
-
Erratum to: Potential of promotion of alleles by genome editing to improve quantitative traits in livestock breeding programs.Genet Sel Evol. 2015 Sep 11;47(1):69. doi: 10.1186/s12711-015-0144-2. Genet Sel Evol. 2015. PMID: 26361936 Free PMC article. No abstract available.
Abstract
Background: Genome editing (GE) is a method that enables specific nucleotides in the genome of an individual to be changed. To date, use of GE in livestock has focussed on simple traits that are controlled by a few quantitative trait nucleotides (QTN) with large effects. The aim of this study was to evaluate the potential of GE to improve quantitative traits that are controlled by many QTN, referred to here as promotion of alleles by genome editing (PAGE).
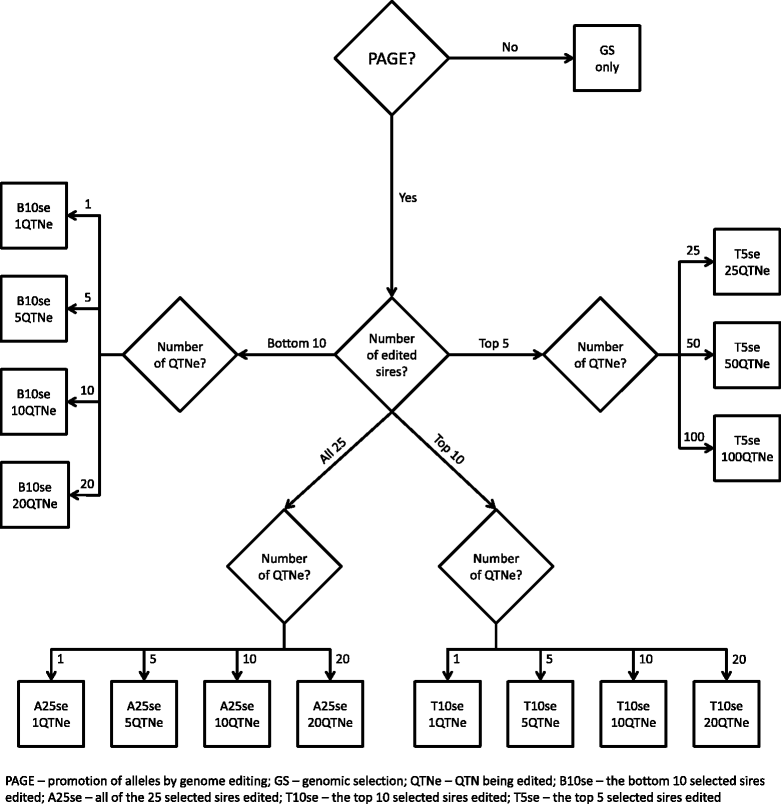
Methods: Multiple scenarios were simulated to test alternative PAGE strategies for a quantitative trait. They differed in (i) the number of edits per sire (0 to 100), (ii) the number of edits per generation (0 to 500), and (iii) the extent of use of PAGE (i.e. editing all sires or only a proportion of them). The base line scenario involved selecting individuals on true breeding values (i.e., genomic selection only (GS only)-genomic selection with perfect accuracy) for several generations. Alternative scenarios complemented this base line scenario with PAGE (GS + PAGE). The effect of different PAGE strategies was quantified by comparing response to selection, changes in allele frequencies, the number of distinct QTN edited, the sum of absolute effects of the edited QTN per generation, and inbreeding.
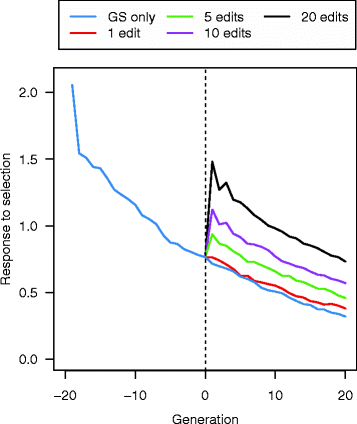
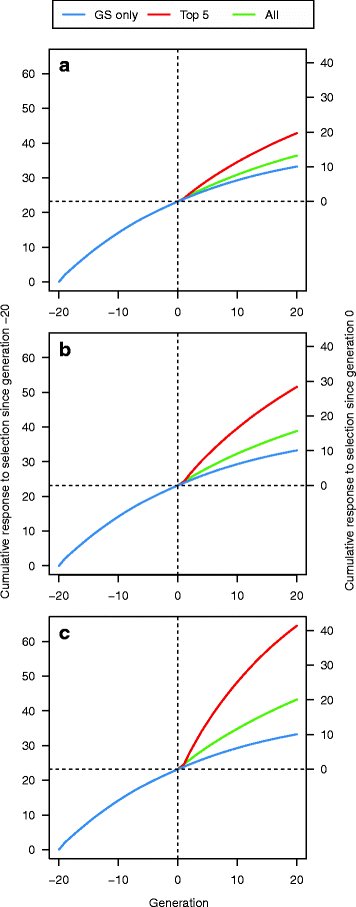
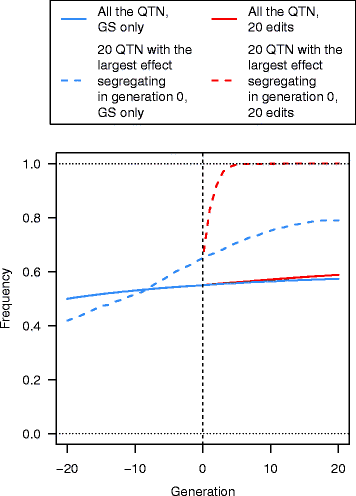
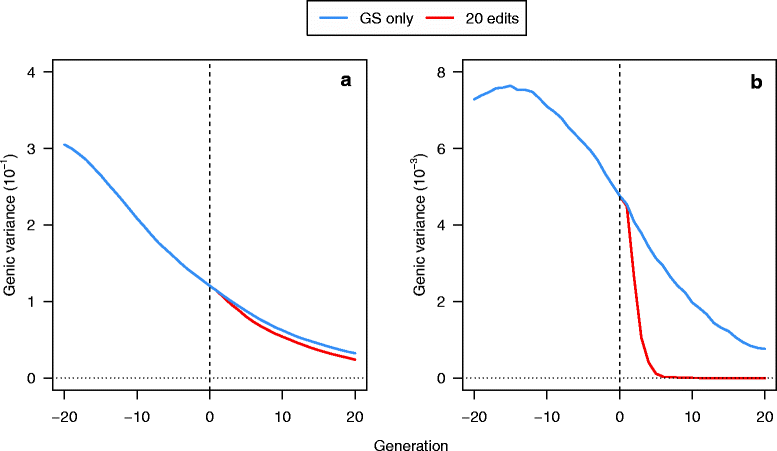
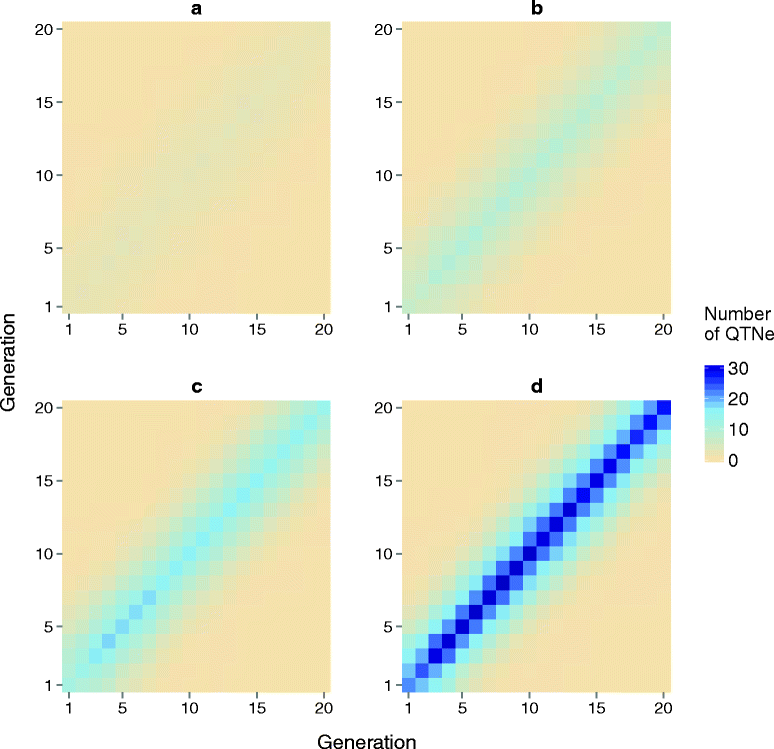
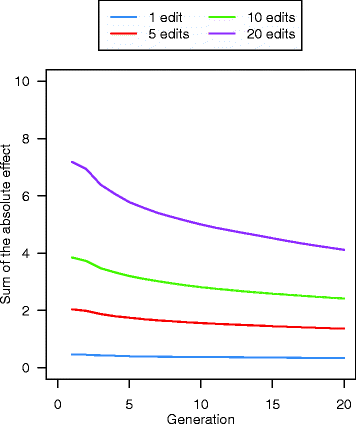
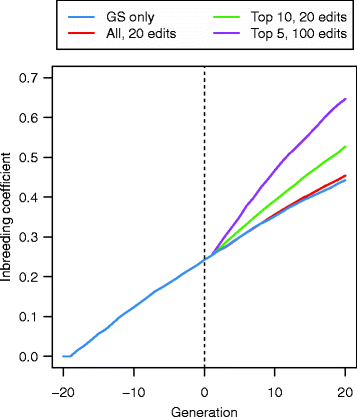
Results: Response to selection after 20 generations was between 1.08 and 4.12 times higher with GS + PAGE than with GS only. Increases in response to selection were larger with more edits per sire and more sires edited. When the total resources for PAGE were limited, editing a few sires for many QTN resulted in greater response to selection and inbreeding compared to editing many sires for a few QTN. Between the scenarios GS only and GS + PAGE, there was little difference in the average change in QTN allele frequencies, but there was a major difference for the QTN with the largest effects. The sum of the effects of the edited QTN decreased across generations.
Conclusions: This study showed that PAGE has great potential for application in livestock breeding programs, but inbreeding needs to be managed.
Figures



References
Publication types
MeSH terms
Grants and funding
- BBS/E/D/05251442/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/D/05251443/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/D/20211553/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MR/M000370/1/MRC_/Medical Research Council/United Kingdom
- BBS/E/D/20211554/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources

