Expression of the Blood-Group-Related Gene B4galnt2 Alters Susceptibility to Salmonella Infection
- PMID: 26133982
- PMCID: PMC4489644
- DOI: 10.1371/journal.ppat.1005008
Expression of the Blood-Group-Related Gene B4galnt2 Alters Susceptibility to Salmonella Infection
Abstract
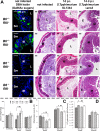
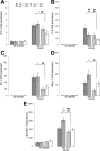
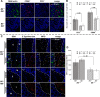
Glycans play important roles in host-microbe interactions. Tissue-specific expression patterns of the blood group glycosyltransferase β-1,4-N-acetylgalactosaminyltransferase 2 (B4galnt2) are variable in wild mouse populations, and loss of B4galnt2 expression is associated with altered intestinal microbiota. We hypothesized that variation in B4galnt2 expression alters susceptibility to intestinal pathogens. To test this, we challenged mice genetically engineered to express different B4galnt2 tissue-specific patterns with a Salmonella Typhimurium infection model. We found B4galnt2 intestinal expression was strongly associated with bacterial community composition and increased Salmonella susceptibility as evidenced by increased intestinal inflammatory cytokines and infiltrating immune cells. Fecal transfer experiments demonstrated a crucial role of the B4galnt2-dependent microbiota in conferring susceptibility to intestinal inflammation, while epithelial B4galnt2 expression facilitated epithelial invasion of S. Typhimurium. These data support a critical role for B4galnt2 in gastrointestinal infections. We speculate that B4galnt2-specific differences in host susceptibility to intestinal pathogens underlie the strong signatures of balancing selection observed at the B4galnt2 locus in wild mouse populations.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






References
-
- Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, Weatherford J, et al. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307(5717):1955–9. - PubMed
-
- Varki A, Freeze HH, Gagneux P. Evolution of Glycan Diversity In: Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, et al. , editors. Essentials of Glycobiology. 2010/03/20 ed. Cold Spring Harbor NY: The Consortium of Glycobiology Editors, La Jolla, California; 2009.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

