The Origin of Mutants Under Selection: How Natural Selection Mimics Mutagenesis (Adaptive Mutation)
- PMID: 26134316
- PMCID: PMC4484973
- DOI: 10.1101/cshperspect.a018176
The Origin of Mutants Under Selection: How Natural Selection Mimics Mutagenesis (Adaptive Mutation)
Abstract
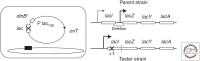
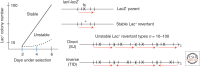
Selection detects mutants but does not cause mutations. Contrary to this dictum, Cairns and Foster plated a leaky lac mutant of Escherichia coli on lactose medium and saw revertant (Lac(+)) colonies accumulate with time above a nongrowing lawn. This result suggested that bacteria might mutagenize their own genome when growth is blocked. However, this conclusion is suspect in the light of recent evidence that revertant colonies are initiated by preexisting cells with multiple copies the conjugative F'lac plasmid, which carries the lac mutation. Some plated cells have multiple copies of the simple F'lac plasmid. This provides sufficient LacZ activity to support plasmid replication but not cell division. In nongrowing cells, repeated plasmid replication increases the likelihood of a reversion event. Reversion to lac(+) triggers exponential cell growth leading to a stable Lac(+) revertant colony. In 10% of these plated cells, the high-copy plasmid includes an internal tandem lac duplication, which provides even more LacZ activity—sufficient to support slow growth and formation of an unstable Lac(+) colony. Cells with multiple copies of the F'lac plasmid have an increased mutation rate, because the plasmid encodes the error-prone (mutagenic) DNA polymerase, DinB. Without DinB, unstable and stable Lac(+) revertant types form in equal numbers and both types arise with no mutagenesis. Amplification and selection are central to behavior of the Cairns-Foster system, whereas mutagenesis is a system-specific side effect or artifact caused by coamplification of dinB with lac. Study of this system has revealed several broadly applicable principles. In all populations, gene duplications are frequent stable genetic polymorphisms, common near-neutral mutant alleles can gain a positive phenotype when amplified under selection, and natural selection can operate without cell division when variability is generated by overreplication of local genome subregions.
Copyright © 2015 Cold Spring Harbor Laboratory Press; all rights reserved.
Figures



Similar articles
-
Selection-Enhanced Mutagenesis of lac Genes Is Due to Their Coamplification with dinB Encoding an Error-Prone DNA Polymerase.Genetics. 2018 Mar;208(3):1009-1021. doi: 10.1534/genetics.117.300409. Epub 2018 Jan 4. Genetics. 2018. PMID: 29301907 Free PMC article.
-
Plasmid copy number underlies adaptive mutability in bacteria.Genetics. 2014 Nov;198(3):919-33. doi: 10.1534/genetics.114.170068. Epub 2014 Aug 29. Genetics. 2014. PMID: 25173846 Free PMC article.
-
Selective Inbreeding: Genetic Crosses Drive Apparent Adaptive Mutation in the Cairns-Foster System of Escherichia coli.Genetics. 2020 Feb;214(2):333-354. doi: 10.1534/genetics.119.302754. Epub 2019 Dec 6. Genetics. 2020. PMID: 31810989 Free PMC article.
-
Adaptive mutation and amplification in Escherichia coli: two pathways of genome adaptation under stress.Res Microbiol. 2004 Jun;155(5):352-9. doi: 10.1016/j.resmic.2004.01.020. Res Microbiol. 2004. PMID: 15207867 Review.
-
Mechanisms of mutation in nondividing cells. Insights from the study of adaptive mutation in Escherichia coli.Ann N Y Acad Sci. 1999 May 18;870:133-45. doi: 10.1111/j.1749-6632.1999.tb08873.x. Ann N Y Acad Sci. 1999. PMID: 10415479 Free PMC article. Review.
Cited by
-
Hopping into a hot seat: Role of DNA structural features on IS5-mediated gene activation and inactivation under stress.PLoS One. 2017 Jun 30;12(6):e0180156. doi: 10.1371/journal.pone.0180156. eCollection 2017. PLoS One. 2017. PMID: 28666002 Free PMC article.
-
A Comprehensive View of Translesion Synthesis in Escherichia coli.Microbiol Mol Biol Rev. 2020 Jun 17;84(3):e00002-20. doi: 10.1128/MMBR.00002-20. Print 2020 Aug 19. Microbiol Mol Biol Rev. 2020. PMID: 32554755 Free PMC article. Review.
-
The sources of adaptive variation.Proc Biol Sci. 2017 May 31;284(1855):20162864. doi: 10.1098/rspb.2016.2864. Proc Biol Sci. 2017. PMID: 28566483 Free PMC article. Review.
-
Inducing stable reversion to achieve cancer control.Nat Rev Cancer. 2016 Apr;16(4):266-70. doi: 10.1038/nrc.2016.12. Nat Rev Cancer. 2016. PMID: 27458638 Review.
-
Natural selection underlies apparent stress-induced mutagenesis in a bacteriophage infection model.Nat Microbiol. 2016 Apr 18;1(6):16047. doi: 10.1038/nmicrobiol.2016.47. Nat Microbiol. 2016. PMID: 27572836
References
-
- Albertson DG. 2006. Gene amplification in cancer. Trends Genet 22: 447–455. - PubMed
-
- Andersson DI, Hughes D, Roth JR. 2011. The origin of mutants under selection: Interactions of mutation, growth, and selection. In EcoSal-Escherichia coli and Salmonella: Cellular and molecular biology (ed. Slauch JM, et al.). ASM, Washington, DC. - PubMed
-
- Bachellier S, Clement JM, Hofnung M. 1999. Short palindromic repetitive DNA elements in enterobacteria: A survey. Res Microbiol 150: 627–639. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
