Comparison of the Oral Microbiomes of Canines and Their Owners Using Next-Generation Sequencing
- PMID: 26134411
- PMCID: PMC4489859
- DOI: 10.1371/journal.pone.0131468
Comparison of the Oral Microbiomes of Canines and Their Owners Using Next-Generation Sequencing
Abstract
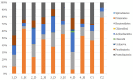
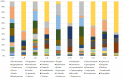
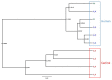
The oral microbiome, which is closely associated with many diseases, and the resident pathogenic oral bacteria, which can be transferred by close physical contact, are important public health considerations. Although the dog is the most common companion animal, the composition of the canine oral microbiome, which may include human pathogenic bacteria, and its relationship with that of their owners are unclear. In this study, 16S rDNA pyrosequencing was used to compare the oral microbiomes of 10 dogs and their owners and to identify zoonotic pathogens. Pyrosequencing revealed 246 operational taxonomic units in the 10 samples, representing 57 genera from eight bacterial phyla. Firmicutes (57.6%), Proteobacteria (21.6%), Bacteroidetes (9.8%), Actinobacteria (7.1%), and Fusobacteria (3.9%) were the predominant phyla in the human oral samples, whereas Proteobacteria (25.7%), Actinobacteria (21%), Bacteroidetes (19.7%), Firmicutes (19.3%), and Fusobacteria (12.3%) were predominant in the canine oral samples. The predominant genera in the human samples were Streptococcus (43.9%), Neisseria (10.3%), Haemophilus (9.6%), Prevotella (8.4%), and Veillonella (8.1%), whereas the predominant genera in the canine samples were Actinomyces (17.2%), Unknown (16.8), Porphyromonas (14.8), Fusobacterium (11.8), and Neisseria (7.2%). The oral microbiomes of dogs and their owners were appreciably different, and similarity in the microbiomes of canines and their owners was not correlated with residing in the same household. Oral-to-oral transfer of Neisseria shayeganii, Porphyromonas canigingivalis, Tannerella forsythia, and Streptococcus minor from dogs to humans was suspected. The finding of potentially zoonotic and periodontopathic bacteria in the canine oral microbiome may be a public health concern.
Conflict of interest statement
Figures

Similar articles
-
Exploring the oral microflora of preschool children.J Microbiol. 2017 Jul;55(7):531-537. doi: 10.1007/s12275-017-6474-8. Epub 2017 Apr 22. J Microbiol. 2017. PMID: 28434085
-
Deep sequencing of the 16S ribosomal RNA of the neonatal oral microbiome: a comparison of breast-fed and formula-fed infants.Sci Rep. 2016 Dec 6;6:38309. doi: 10.1038/srep38309. Sci Rep. 2016. PMID: 27922070 Free PMC article.
-
Highly diverse microbiota in dental root canals in cases of apical periodontitis (data of illumina sequencing).J Endod. 2014 Nov;40(11):1778-83. doi: 10.1016/j.joen.2014.06.017. Epub 2014 Sep 16. J Endod. 2014. PMID: 25227214
-
Gut microbiota of humans, dogs and cats: current knowledge and future opportunities and challenges.Br J Nutr. 2015 Jan;113 Suppl:S6-17. doi: 10.1017/S0007114514002943. Epub 2014 Nov 21. Br J Nutr. 2015. PMID: 25414978 Review.
-
Current state of knowledge: the canine gastrointestinal microbiome.Anim Health Res Rev. 2012 Jun;13(1):78-88. doi: 10.1017/S1466252312000059. Epub 2012 May 30. Anim Health Res Rev. 2012. PMID: 22647637 Review.
Cited by
-
The Environmental Microbiome, Allergic Disease, and Asthma.J Allergy Clin Immunol Pract. 2022 Sep;10(9):2206-2217.e1. doi: 10.1016/j.jaip.2022.06.006. Epub 2022 Jun 22. J Allergy Clin Immunol Pract. 2022. PMID: 35750322 Free PMC article.
-
Identification of bacteria associated with canine otitis externa based on 16S rDNA high-throughput sequencing.Braz J Microbiol. 2023 Dec;54(4):3283-3290. doi: 10.1007/s42770-023-01166-0. Epub 2023 Oct 27. Braz J Microbiol. 2023. PMID: 37889464 Free PMC article.
-
Genomics of the Uncultivated, Periodontitis-Associated Bacterium Tannerella sp. BU045 (Oral Taxon 808).mSystems. 2018 Jun 5;3(3):e00018-18. doi: 10.1128/mSystems.00018-18. eCollection 2018 May-Jun. mSystems. 2018. PMID: 29896567 Free PMC article.
-
Optimizing methods and dodging pitfalls in microbiome research.Microbiome. 2017 May 5;5(1):52. doi: 10.1186/s40168-017-0267-5. Microbiome. 2017. PMID: 28476139 Free PMC article. Review.
-
Canibacter oris - a fairly unknown pathogenic agent of bite wound infections.GMS Infect Dis. 2021 Apr 22;9:Doc01. doi: 10.3205/id000070. eCollection 2021. GMS Infect Dis. 2021. PMID: 34113533 Free PMC article.
References
-
- Fowler EB, Breault LG, Cuenin MF. Periodontal disease and its association with systemic disease. Mil Med. 2001;166: 85–89. - PubMed
-
- Darby I, Curtis M. Microbiology of periodontal disease in children and young adults. Periodontol 2000. 2001;26: 33–53. Available: http://www.ncbi.nlm.nih.gov/pubmed/11452905 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

