Identification of Alp1U and Lom6 as epoxy hydrolases and implications for kinamycin and lomaiviticin biosynthesis
- PMID: 26134788
- PMCID: PMC4506494
- DOI: 10.1038/ncomms8674
Identification of Alp1U and Lom6 as epoxy hydrolases and implications for kinamycin and lomaiviticin biosynthesis
Abstract
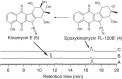
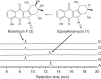
The naturally occurring diazobenzofluorenes, kinamycins, fluostatins and lomaiviticins, possess highly oxygenated A-rings, via which the last forms a dimeric pharmacophore. However, neither the A-ring transformation nor the dimerization mechanisms have been explored thus far. Here we propose a unified biosynthetic logic for the three types of antibiotics and verify one key reaction via detailed genetic and enzymatic experiments. Alp1U and Lom6 from the kinamycin and lomaiviticin biosynthesis, respectively, are shown to catalyse epoxy hydrolysis on a substrate that is obtained by chemical deacetylation of a kinamycin-pathway-derived intermediate. Thus, our study provides the first evidence for the existence of an epoxy intermediate in lomaiviticin biosynthesis. Furthermore, our results suggest that the dimerization in the lomaiviticin biosynthesis proceeds after dehydration of a product generated by Lom6.
Figures


Similar articles
-
Mutation of an atypical oxirane oxyanion hole improves regioselectivity of the α/β-fold epoxide hydrolase Alp1U.J Biol Chem. 2020 Dec 11;295(50):16987-16997. doi: 10.1074/jbc.RA120.015563. Epub 2020 Oct 1. J Biol Chem. 2020. PMID: 33004437 Free PMC article.
-
Insights into lomaiviticin biosynthesis. Isolation and structure elucidation of (-)-homoseongomycin.J Nat Prod. 2013 Jul 26;76(7):1238-41. doi: 10.1021/np400355h. Epub 2013 Jun 26. J Nat Prod. 2013. PMID: 23803003
-
Predictive model for epoxide hydrolase-generated stereochemistry in the biosynthesis of nine-membered enediyne antitumor antibiotics.Biochemistry. 2013 Aug 6;52(31):5217-24. doi: 10.1021/bi400572a. Epub 2013 Jul 23. Biochemistry. 2013. PMID: 23844627 Free PMC article.
-
The diazofluorene antitumor antibiotics: structural elucidation, biosynthetic, synthetic, and chemical biological studies.Nat Prod Rep. 2012 Jan;29(1):87-118. doi: 10.1039/c1np00052g. Epub 2011 Oct 28. Nat Prod Rep. 2012. PMID: 22037715 Review.
-
New structural and chemical insight into the catalytic mechanism of epoxide hydrolases.Drug Metab Rev. 2000 Aug-Nov;32(3-4):327-38. doi: 10.1081/dmr-100102337. Drug Metab Rev. 2000. PMID: 11139132 Review.
Cited by
-
Biochemical and structural insights of multifunctional flavin-dependent monooxygenase FlsO1-catalyzed unexpected xanthone formation.Nat Commun. 2022 Sep 14;13(1):5386. doi: 10.1038/s41467-022-33131-0. Nat Commun. 2022. PMID: 36104338 Free PMC article.
-
The functional differentiation of the post-PKS tailoring oxygenases contributed to the chemical diversities of atypical angucyclines.Synth Syst Biotechnol. 2018 Nov 14;3(4):275-282. doi: 10.1016/j.synbio.2018.11.001. eCollection 2018 Dec. Synth Syst Biotechnol. 2018. PMID: 30533539 Free PMC article. Review.
-
Use of genome-scale models to get new insights into the marine actinomycete genus Salinispora.BMC Syst Biol. 2019 Jan 21;13(1):11. doi: 10.1186/s12918-019-0683-1. BMC Syst Biol. 2019. PMID: 30665399 Free PMC article.
-
Molecular Dynamics to Elucidate the DNA-Binding Activity of AlpZ, a Member of the Gamma-Butyrolactone Receptor Family in Streptomyces ambofaciens.Front Microbiol. 2020 Jul 2;11:1255. doi: 10.3389/fmicb.2020.01255. eCollection 2020. Front Microbiol. 2020. PMID: 32714286 Free PMC article.
-
Mutation of an atypical oxirane oxyanion hole improves regioselectivity of the α/β-fold epoxide hydrolase Alp1U.J Biol Chem. 2020 Dec 11;295(50):16987-16997. doi: 10.1074/jbc.RA120.015563. Epub 2020 Oct 1. J Biol Chem. 2020. PMID: 33004437 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

