Hepadnavirus Genome Replication and Persistence
- PMID: 26134841
- PMCID: PMC4484952
- DOI: 10.1101/cshperspect.a021386
Hepadnavirus Genome Replication and Persistence
Abstract
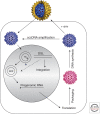
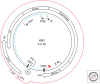
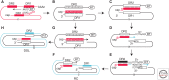
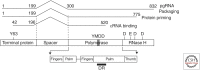
Hallmarks of the hepadnavirus replication cycle are the formation of covalently closed circular DNA (cccDNA) and the reverse transcription of a pregenomic RNA (pgRNA) in core particles leading to synthesis of the relaxed circular DNA (rcDNA) genome. cccDNA, the template for viral RNA transcription, is the basis for the persistence of these viruses in infected hepatocytes. In this review, we summarize the current state of knowledge on the mechanisms of hepadnavirus reverse transcription and the biochemical and structural properties of the viral reverse transcriptase (RT). We highlight important gaps in knowledge regarding cccDNA biosynthesis and stability. In addition, we discuss the impact of current antiviral therapies on viral persistence, particularly on cccDNA.
Copyright © 2015 Cold Spring Harbor Laboratory Press; all rights reserved.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
