Revealing microbial recognition by specific antibodies
- PMID: 26134992
- PMCID: PMC4489363
- DOI: 10.1186/s12866-015-0456-y
Revealing microbial recognition by specific antibodies
Abstract
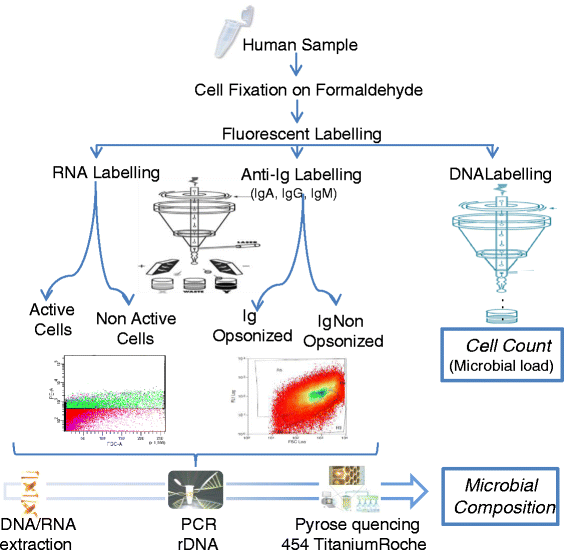
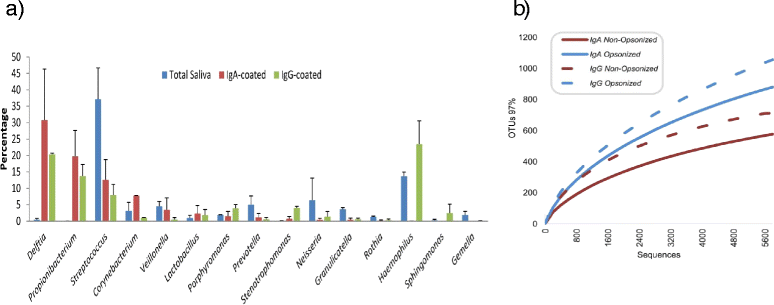
Background: Recognition of microorganisms by antibodies is a vital component of the human immune response. However, there is currently very limited understanding of immune recognition of 50 % of the human microbiome which is made up of as yet un-culturable bacteria. We have combined the use of flow cytometry and pyrosequencing to describe the microbial composition of human samples, and its interaction with the immune system.
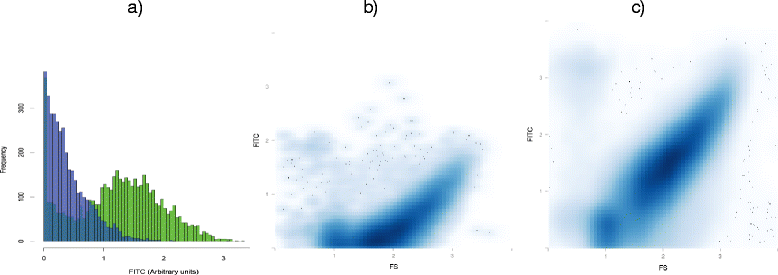
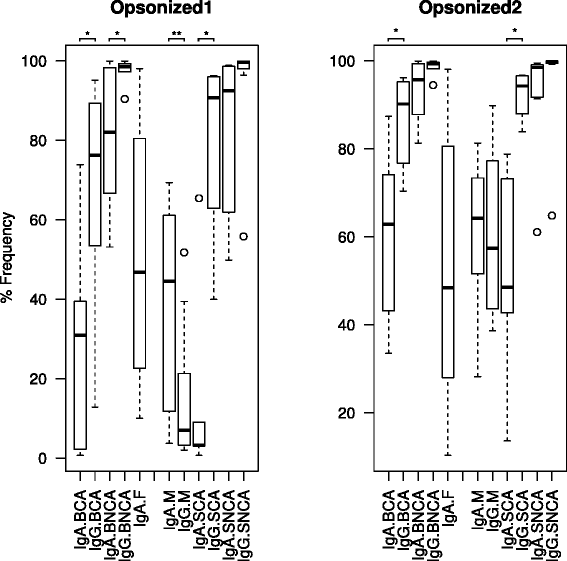
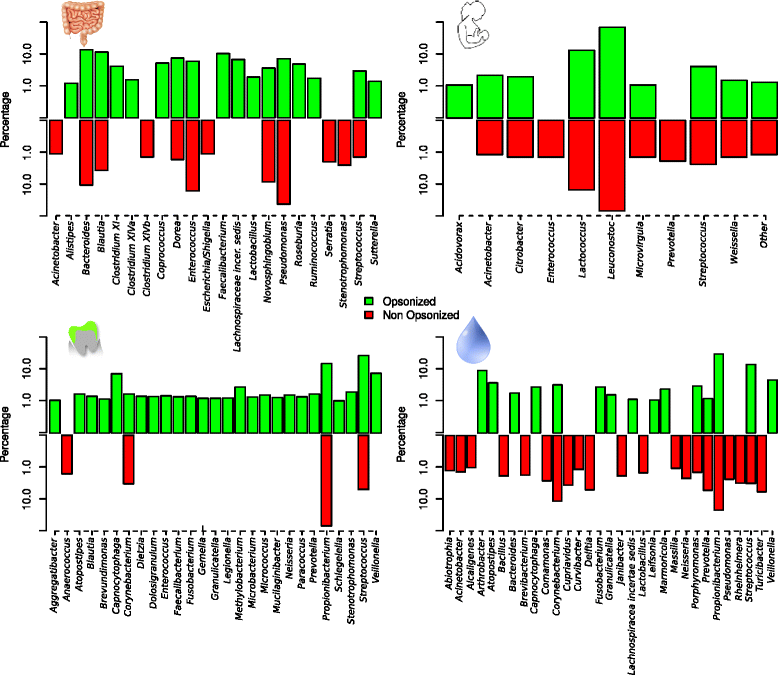
Results: We show the power of the technique in human faecal, saliva, oral biofilm and breast milk samples, labeled with fluorescent anti-IgG or anti-IgA antibodies. Using Fluorescence-Activated Cell Sorting (FACS), bacterial cells were separated depending on whether they are coated with IgA or IgG antibodies. Each bacterial population was PCR-amplified and pyrosequenced, characterizing the microorganisms which evade the immune system and those which were recognized by each immunoglobulin.
Conclusions: The application of the technique to healthy and diseased individuals may unravel the contribution of the immune response to microbial infections and polymicrobial diseases.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

