Defining and Evaluating a Core Genome Multilocus Sequence Typing Scheme for Whole-Genome Sequence-Based Typing of Listeria monocytogenes
- PMID: 26135865
- PMCID: PMC4540939
- DOI: 10.1128/JCM.01193-15
Defining and Evaluating a Core Genome Multilocus Sequence Typing Scheme for Whole-Genome Sequence-Based Typing of Listeria monocytogenes
Abstract
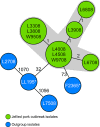
Whole-genome sequencing (WGS) has emerged today as an ultimate typing tool to characterize Listeria monocytogenes outbreaks. However, data analysis and interlaboratory comparability of WGS data are still challenging for most public health laboratories. Therefore, we have developed and evaluated a new L. monocytogenes typing scheme based on genome-wide gene-by-gene comparisons (core genome multilocus the sequence typing [cgMLST]) to allow for a unique typing nomenclature. Initially, we determined the breadth of the L. monocytogenes population based on MLST data with a Bayesian approach. Based on the genome sequence data of representative isolates for the whole population, cgMLST target genes were defined and reappraised with 67 L. monocytogenes isolates from two outbreaks and serotype reference strains. The Bayesian population analysis generated five L. monocytogenes groups. Using all available NCBI RefSeq genomes (n = 36) and six additionally sequenced strains, all genetic groups were covered. Pairwise comparisons of these 42 genome sequences resulted in 1,701 cgMLST targets present in all 42 genomes with 100% overlap and ≥90% sequence similarity. Overall, ≥99.1% of the cgMLST targets were present in 67 outbreak and serotype reference strains, underlining the representativeness of the cgMLST scheme. Moreover, cgMLST enabled clustering of outbreak isolates with ≤10 alleles difference and unambiguous separation from unrelated outgroup isolates. In conclusion, the novel cgMLST scheme not only improves outbreak investigations but also enables, due to the availability of the automatically curated cgMLST nomenclature, interlaboratory exchange of data that are crucial, especially for rapid responses during transsectorial outbreaks.
Copyright © 2015 Ruppitsch et al.
Figures

References
-
- Allerberger F, Bagó Z, Huhulescu S, Pietzka A. 2015. Listeriosis: the dark side of refrigeration and ensiling, p 249–286. In Sing A. (ed), Zoonoses—infections affecting humans and animals. focus on public health aspects Springer Verlag, Heidelberg, Germany.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

