Exploring the Balance between DNA Pressure and Capsid Stability in Herpesviruses and Phages
- PMID: 26136570
- PMCID: PMC4542358
- DOI: 10.1128/JVI.01172-15
Exploring the Balance between DNA Pressure and Capsid Stability in Herpesviruses and Phages
Abstract
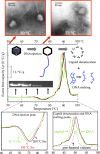
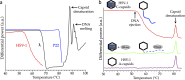
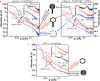
We have recently shown in both herpesviruses and phages that packaged viral DNA creates a pressure of tens of atmospheres pushing against the interior capsid wall. For the first time, using differential scanning microcalorimetry, we directly measured the energy powering the release of pressurized DNA from the capsid. Furthermore, using a new calorimetric assay to accurately determine the temperature inducing DNA release, we found a direct influence of internal DNA pressure on the stability of the viral particle. We show that the balance of forces between the DNA pressure and capsid strength, required for DNA retention between rounds of infection, is conserved between evolutionarily diverse bacterial viruses (phages λ and P22), as well as a eukaryotic virus, human herpes simplex 1 (HSV-1). Our data also suggest that the portal vertex in these viruses is the weakest point in the overall capsid structure and presents the Achilles heel of the virus's stability. Comparison between these viral systems shows that viruses with higher DNA packing density (resulting in higher capsid pressure) have inherently stronger capsid structures, preventing spontaneous genome release prior to infection. This force balance is of key importance for viral survival and replication. Investigating the ways to disrupt this balance can lead to development of new mutation-resistant antivirals.
Importance: A virus can generally be described as a nucleic acid genome contained within a protective protein shell, called the capsid. For many double-stranded DNA viruses, confinement of the large DNA molecule within the small protein capsid results in an energetically stressed DNA state exerting tens of atmospheres of pressures on the inner capsid wall. We show that stability of viral particles (which directly relates to infectivity) is strongly influenced by the state of the packaged genome. Using scanning calorimetry on a bacterial virus (phage λ) as an experimental model system, we investigated the thermodynamics of genome release associated with destabilizing the viral particle. Furthermore, we compare the influence of tight genome confinement on the relative stability for diverse bacterial and eukaryotic viruses. These comparisons reveal an evolutionarily conserved force balance between the capsid stability and the density of the packaged genome.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures




References
-
- Flint SJ. 2009. Principles of virology, 3rd ed ASM Press, Washington, DC.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

