The complete chloroplast genome sequence of the relict woody plant Metasequoia glyptostroboides Hu et Cheng
- PMID: 26136762
- PMCID: PMC4468836
- DOI: 10.3389/fpls.2015.00447
The complete chloroplast genome sequence of the relict woody plant Metasequoia glyptostroboides Hu et Cheng
Abstract
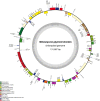
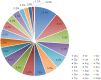
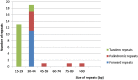
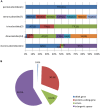
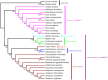
Metasequoia glyptostroboides Hu et Cheng is the only species in the genus Metasequoia Miki ex Hu et Cheng, which belongs to the Cupressaceae family. There were around 10 species in the Metasequoia genus, which were widely spread across the Northern Hemisphere during the Cretaceous of the Mesozoic and in the Cenozoic. M. glyptostroboides is the only remaining representative of this genus. Here, we report the complete chloroplast (cp) genome sequence and the cp genomic features of M. glyptostroboides. The M. glyptostroboides cp genome is 131,887 bp in length, with a total of 117 genes comprised of 82 protein-coding genes, 31 tRNA genes and four rRNA genes. In this genome, 11 forward repeats, nine palindromic repeats, and 15 tandem repeats were detected. A total of 188 perfect microsatellites were detected through simple sequence repeat (SSR) analysis and these were distributed unevenly within the cp genome. Comparison of the cp genome structure and gene order to those of several other land plants indicated that a copy of the inverted repeat (IR) region, which was found to be IR region A (IRA), was lost in the M. glyptostroboides cp genome. The five most divergent and five most conserved genes were determined and further phylogenetic analysis was performed among plant species, especially for related species in conifers. Finally, phylogenetic analysis demonstrated that M. glyptostroboides is a sister species to Cryptomeria japonica (L. F.) D. Don and to Taiwania cryptomerioides Hayata. The complete cp genome sequence information of M. glyptostroboides will be great helpful for further investigations of this endemic relict woody plant and for in-depth understanding of the evolutionary history of the coniferous cp genomes, especially for the position of M. glyptostroboides in plant systematics and evolution.
Keywords: Metasequoia glyptostroboides; chloroplast genome; conifer evolution; cupressophytes; inverted repeat; relict plant.
Figures
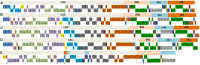
References
-
- Bartholomew B., Boufford D. E., Spongberg S. A. (1983). Metasequoia glyptostroboides-Its present status in central China. J. Arnold Arbor. 64 105–128.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

