Cell-selective labelling of proteomes in Drosophila melanogaster
- PMID: 26138272
- PMCID: PMC4507001
- DOI: 10.1038/ncomms8521
Cell-selective labelling of proteomes in Drosophila melanogaster
Abstract
The specification and adaptability of cells rely on changes in protein composition. Nonetheless, uncovering proteome dynamics with cell-type-specific resolution remains challenging. Here we introduce a strategy for cell-specific analysis of newly synthesized proteomes by combining targeted expression of a mutated methionyl-tRNA synthetase (MetRS) with bioorthogonal or fluorescent non-canonical amino-acid-tagging techniques (BONCAT or FUNCAT). Substituting leucine by glycine within the MetRS-binding pocket (MetRS(LtoG)) enables incorporation of the non-canonical amino acid azidonorleucine (ANL) instead of methionine during translation. Newly synthesized proteins can thus be labelled by coupling the azide group of ANL to alkyne-bearing tags through 'click chemistry'. To test these methods for applicability in vivo, we expressed MetRS(LtoG) cell specifically in Drosophila. FUNCAT and BONCAT reveal ANL incorporation into proteins selectively in cells expressing the mutated enzyme. Cell-type-specific FUNCAT and BONCAT, thus, constitute eligible techniques to study protein synthesis-dependent processes in complex and behaving organisms.
Figures
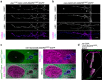


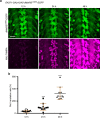
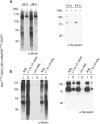
References
-
- Pandey A. & Mann M. Proteomics to study genes and genomes. Nature 405, 837–846 (2000). - PubMed
-
- Burkhart J. M. et al. The first comprehensive and quantitative analysis of human platelet protein composition allows the comparative analysis of structural and functional pathways. Blood 120, e73–e82 (2012). - PubMed
-
- Wasbrough E. R. et al. The Drosophila melanogaster sperm proteome-II (DmSP-II). J. Proteomics 73, 2171–2185 (2010). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

