SMAP: a streamlined methylation analysis pipeline for bisulfite sequencing
- PMID: 26140213
- PMCID: PMC4488126
- DOI: 10.1186/s13742-015-0070-9
SMAP: a streamlined methylation analysis pipeline for bisulfite sequencing
Abstract
Background: DNA methylation has important roles in the regulation of gene expression and cellular specification. Reduced representation bisulfite sequencing (RRBS) has prevailed in methylation studies due to its cost-effectiveness and single-base resolution. The rapid accumulation of RRBS data demands well designed analytical tools.
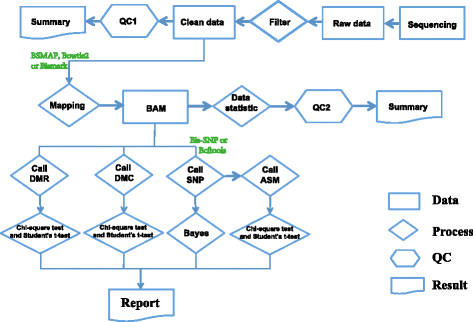
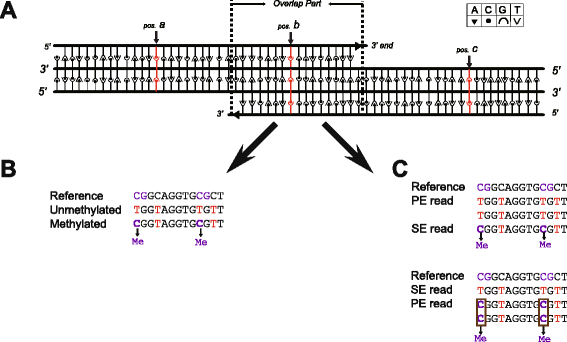
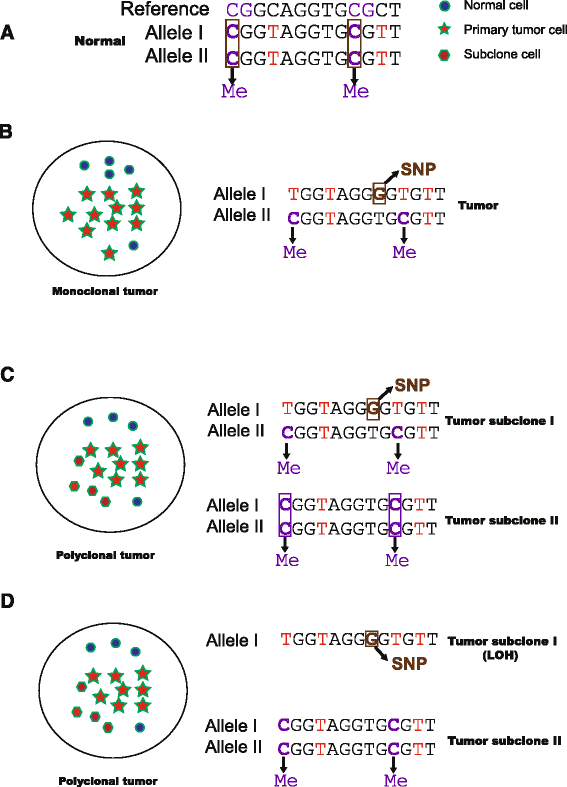
Findings: To streamline the data processing of DNA methylation from multiple RRBS samples, we present a flexible pipeline named SMAP, whose features include: (i) handling of single-and/or paired-end diverse bisulfite sequencing data with reduced false-positive rates in differentially methylated regions; (ii) detection of allele-specific methylation events with improved algorithms; (iii) a built-in pipeline for detection of novel single nucleotide polymorphisms (SNPs); (iv) support of multiple user-defined restriction enzymes; (v) conduction of all methylation analyses in a single-step operation when well configured.
Conclusions: Simulation and experimental data validated the high accuracy of SMAP for SNP detection and methylation identification. Most analyses required in methylation studies (such as estimation of methylation levels, differentially methylated cytosine groups, and allele-specific methylation regions) can be executed readily with SMAP. All raw data from diverse samples could be processed in parallel and 'packetized' streams. A simple user guide to the methylation applications is also provided.
Keywords: Allele-specific DNA methylation (ASM); Differentially methylated region (DMR); Reduced representation bisulfite sequencing (RRBS).
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

