Histone H2B Monoubiquitination Mediated by HISTONE MONOUBIQUITINATION1 and HISTONE MONOUBIQUITINATION2 Is Involved in Anther Development by Regulating Tapetum Degradation-Related Genes in Rice
- PMID: 26143250
- PMCID: PMC4528728
- DOI: 10.1104/pp.114.256578
Histone H2B Monoubiquitination Mediated by HISTONE MONOUBIQUITINATION1 and HISTONE MONOUBIQUITINATION2 Is Involved in Anther Development by Regulating Tapetum Degradation-Related Genes in Rice
Abstract
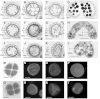
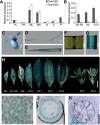
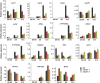
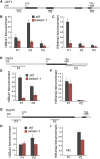
Histone H2B monoubiquitination (H2Bub1) is an important regulatory mechanism in eukaryotic gene transcription and is essential for normal plant development. However, the function of H2Bub1 in reproductive development remains elusive. Here, we report rice (Oryza sativa) HISTONE MONOUBIQUITINATION1 (OsHUB1) and OsHUB2, the homologs of Arabidopsis (Arabidopsis thaliana) HUB1 and HUB2 proteins, which function as E3 ligases in H2Bub1, are involved in late anther development in rice. oshub mutants exhibit abnormal tapetum development and aborted pollen in postmeiotic anthers. Knockout of OsHUB1 or OsHUB2 results in the loss of H2Bub1 and a reduction in the levels of dimethylated lysine-4 on histone 3 (H3K4me2). Anther transcriptome analysis revealed that several key tapetum degradation-related genes including OsC4, rice Cysteine Protease1 (OsCP1), and Undeveloped Tapetum1 (UDT1) were down-regulated in the mutants. Further, chromatin immunoprecipitation assays demonstrate that H2Bub1 directly targets OsC4, OsCP1, and UDT1 genes, and enrichment of H2Bub1 and H3K4me2 in the targets is consistent to some degree. Our studies suggest that histone H2B monoubiquitination, mediated by OsHUB1 and OsHUB2, is an important epigenetic modification that in concert with H3K4me2, modulates transcriptional regulation of anther development in rice.
© 2015 American Society of Plant Biologists. All Rights Reserved.
Figures





References
-
- Berr A, Shafiq S, Shen WH (2011) Histone modifications in transcriptional activation during plant development. Biochim Biophys Acta 1809: 567–576 - PubMed
-
- Bowler C, Benvenuto G, Laflamme P, Molino D, Probst AV, Tariq M, Paszkowski J (2004) Chromatin techniques for plant cells. Plant J 39: 776–789 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

