Multidimensional High-Resolution Magic Angle Spinning and Solution-State NMR Characterization of (13)C-labeled Plant Metabolites and Lignocellulose
- PMID: 26143886
- PMCID: PMC4491710
- DOI: 10.1038/srep11848
Multidimensional High-Resolution Magic Angle Spinning and Solution-State NMR Characterization of (13)C-labeled Plant Metabolites and Lignocellulose
Abstract
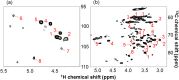
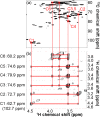
Lignocellulose, which includes mainly cellulose, hemicellulose, and lignin, is a potential resource for the production of chemicals and for other applications. For effective production of materials derived from biomass, it is important to characterize the metabolites and polymeric components of the biomass. Nuclear magnetic resonance (NMR) spectroscopy has been used to identify biomass components; however, the NMR spectra of metabolites and lignocellulose components are ambiguously assigned in many cases due to overlapping chemical shift peaks. Using our (13)C-labeling technique in higher plants such as poplar samples, we demonstrated that overlapping peaks could be resolved by three-dimensional NMR experiments to more accurately assign chemical shifts compared with two-dimensional NMR measurements. Metabolites of the (13)C-poplar were measured by high-resolution magic angle spinning NMR spectroscopy, which allows sample analysis without solvent extraction, while lignocellulose components of the (13)C-poplar dissolved in dimethylsulfoxide/pyridine solvent were analyzed by solution-state NMR techniques. Using these methods, we were able to unambiguously assign chemical shifts of small and macromolecular components in (13)C-poplar samples. Furthermore, using samples of less than 5 mg, we could differentiate between two kinds of genes that were overexpressed in poplar samples, which produced clearly modified plant cell wall components.
Figures




Similar articles
-
Solution-state 2D NMR spectroscopy of plant cell walls enabled by a dimethylsulfoxide-d6/1-ethyl-3-methylimidazolium acetate solvent.Anal Chem. 2013 Mar 19;85(6):3213-21. doi: 10.1021/ac303529v. Epub 2013 Mar 4. Anal Chem. 2013. PMID: 23413964
-
Solution-state 2D NMR of ball-milled plant cell wall gels in DMSO-d(6)/pyridine-d(5).Org Biomol Chem. 2010 Feb 7;8(3):576-91. doi: 10.1039/b916070a. Epub 2009 Dec 3. Org Biomol Chem. 2010. PMID: 20090974 Free PMC article.
-
Comprehensive signal assignment of 13C-labeled lignocellulose using multidimensional solution NMR and 13C chemical shift comparison with solid-state NMR.Anal Chem. 2013 Sep 17;85(18):8857-65. doi: 10.1021/ac402197h. Epub 2013 Sep 6. Anal Chem. 2013. PMID: 24010724
-
Advances in solid-state NMR of cellulose.Curr Opin Biotechnol. 2014 Jun;27:176-84. doi: 10.1016/j.copbio.2014.02.002. Epub 2014 Mar 2. Curr Opin Biotechnol. 2014. PMID: 24590189 Review.
-
Multidimensional solid-state NMR spectroscopy of plant cell walls.Solid State Nucl Magn Reson. 2016 Sep;78:56-63. doi: 10.1016/j.ssnmr.2016.08.001. Epub 2016 Aug 13. Solid State Nucl Magn Reson. 2016. PMID: 27552739 Free PMC article. Review.
Cited by
-
Metabolic Profiling of Intact Arabidopsis thaliana Leaves during Circadian Cycle Using 1H High Resolution Magic Angle Spinning NMR.PLoS One. 2016 Sep 23;11(9):e0163258. doi: 10.1371/journal.pone.0163258. eCollection 2016. PLoS One. 2016. PMID: 27662620 Free PMC article.
-
Tetraploidization promotes radial stem growth in poplars.Plant Biotechnol (Tokyo). 2022 Sep 25;39(3):215-220. doi: 10.5511/plantbiotechnology.22.0716a. Plant Biotechnol (Tokyo). 2022. PMID: 36349238 Free PMC article.
-
InterSpin: Integrated Supportive Webtools for Low- and High-Field NMR Analyses Toward Molecular Complexity.ACS Omega. 2019 Feb 14;4(2):3361-3369. doi: 10.1021/acsomega.8b02714. eCollection 2019 Feb 28. ACS Omega. 2019. PMID: 31459550 Free PMC article.
-
Signal Deconvolution and Generative Topographic Mapping Regression for Solid-State NMR of Multi-Component Materials.Int J Mol Sci. 2021 Jan 22;22(3):1086. doi: 10.3390/ijms22031086. Int J Mol Sci. 2021. PMID: 33499371 Free PMC article.
-
Enhancement of Secondary Cell Wall Formation in Poplar Xylem Using a Self-Reinforced System of Secondary Cell Wall-Related Transcription Factors.Front Plant Sci. 2022 Mar 14;13:819360. doi: 10.3389/fpls.2022.819360. eCollection 2022. Front Plant Sci. 2022. PMID: 35371169 Free PMC article.
References
-
- Tilman D., Hill J. & Lehman C. Carbon-negative biofuels from low-input high-diversity grassland biomass. Science 314, 1598–1600 (2006). - PubMed
-
- Sticklen M. B. Plant genetic engineering for biofuel production: towards affordable cellulosic ethanol. Nat. Rev. Genet. 9, 433–443 (2008). - PubMed
-
- Rubin E. M. Genomics of cellulosic biofuels. Nature 454, 841–845 (2008). - PubMed
-
- Noor E., Eden E., Milo R. & Alon U. Central carbon metabolism as a minimal biochemical walk between precursors for biomass and energy. Mol. Cell 39, 809–820 (2010). - PubMed
-
- Lee C., Teng Q., Zhong R. & Ye Z. H. The four Arabidopsis reduced wall acetylation genes are expressed in secondary wall-containing cells and required for the acetylation of xylan. Plant Cell Physiol. 52, 1289–1301 (2011). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

