Suppression of the alternative lengthening of telomere pathway by the chromatin remodelling factor ATRX
- PMID: 26143912
- PMCID: PMC4501375
- DOI: 10.1038/ncomms8538
Suppression of the alternative lengthening of telomere pathway by the chromatin remodelling factor ATRX
Abstract
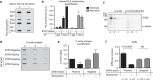
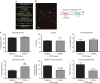
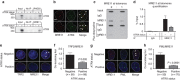
Fifteen per cent of cancers maintain telomere length independently of telomerase by the homologous recombination (HR)-associated alternative lengthening of telomeres (ALT) pathway. A unifying feature of these tumours are mutations in ATRX. Here we show that expression of ectopic ATRX triggers a suppression of the pathway and telomere shortening. Importantly ATRX-mediated ALT suppression is dependent on the histone chaperone DAXX. Re-expression of ATRX is associated with a reduction in replication fork stalling, a known trigger for HR and loss of MRN from telomeres. A G-quadruplex stabilizer partially reverses the effect of ATRX, inferring ATRX may normally facilitate replication through these sequences that, if they persist, promote ALT. We propose that defective telomere chromatinization through loss of ATRX promotes the persistence of aberrant DNA secondary structures, which in turn present a barrier to DNA replication, leading to replication fork stalling, collapse, HR and subsequent recombination-mediated telomere synthesis in ALT cancers.
Figures



References
-
- Gilson E. & Geli V. How telomeres are replicated. Nat. Rev. Mol. Cell Biol. 8, 825–838 (2007). - PubMed
-
- Bryan T. M., Englezou A., Dalla-Pozza L., Dunham M. A. & Reddel R. R. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat. Med. 3, 1271–1274 (1997). - PubMed
-
- Dunham M. A., Neumann A. A., Fasching C. L. & Reddel R. R. Telomere maintenance by recombination in human cells. Nat. Genet. 26, 447–450 (2000). - PubMed
-
- Perrem K. et al.. Repression of an alternative mechanism for lengthening of telomeres in somatic cell hybrids. Oncogene 18, 3383–3390 (1999). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

