The circadian clock rephases during lateral root organ initiation in Arabidopsis thaliana
- PMID: 26144255
- PMCID: PMC4506504
- DOI: 10.1038/ncomms8641
The circadian clock rephases during lateral root organ initiation in Arabidopsis thaliana
Abstract
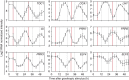
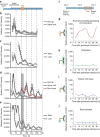
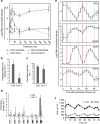
The endogenous circadian clock enables organisms to adapt their growth and development to environmental changes. Here we describe how the circadian clock is employed to coordinate responses to the key signal auxin during lateral root (LR) emergence. In the model plant, Arabidopsis thaliana, LRs originate from a group of stem cells deep within the root, necessitating that new organs emerge through overlying root tissues. We report that the circadian clock is rephased during LR development. Metabolite and transcript profiling revealed that the circadian clock controls the levels of auxin and auxin-related genes including the auxin response repressor IAA14 and auxin oxidase AtDAO2. Plants lacking or overexpressing core clock components exhibit LR emergence defects. We conclude that the circadian clock acts to gate auxin signalling during LR development to facilitate organ emergence.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
- BB/F005318/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/K018078/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/G023972/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/F005237/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/D017904/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

