Universal allosteric mechanism for Gα activation by GPCRs
- PMID: 26147082
- PMCID: PMC4866443
- DOI: 10.1038/nature14663
Universal allosteric mechanism for Gα activation by GPCRs
Abstract
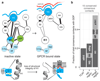
G protein-coupled receptors (GPCRs) allosterically activate heterotrimeric G proteins and trigger GDP release. Given that there are ∼800 human GPCRs and 16 different Gα genes, this raises the question of whether a universal allosteric mechanism governs Gα activation. Here we show that different GPCRs interact with and activate Gα proteins through a highly conserved mechanism. Comparison of Gα with the small G protein Ras reveals how the evolution of short segments that undergo disorder-to-order transitions can decouple regions important for allosteric activation from receptor binding specificity. This might explain how the GPCR-Gα system diversified rapidly, while conserving the allosteric activation mechanism.
Figures