Lhx6 and Lhx8: cell fate regulators and beyond
- PMID: 26148970
- PMCID: PMC4566936
- DOI: 10.1096/fj.14-267500
Lhx6 and Lhx8: cell fate regulators and beyond
Abstract
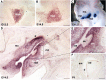
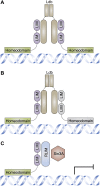
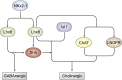
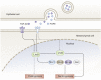
As transcription factors of the lines (LIN)-11/Islet (Isl)-1/mitosis entry checkpoint (MEC)-3 (LIM)-homeobox subfamily, LIM homeobox (Lhx)6 and -8 are remarkably conserved and involved in the morphogenesis of multiple organ systems. Lhx6 and -8 play overlapping and distinctive roles, but in general act as cell fate mediators and in turn are regulated by several transcriptional factors, such as sonic hedgehog, fibroblast growth factors, and wingless-int (Wnt)/β-catenin. In this review, we first summarize Lhx6 and -8 distributions in development and then explore how Lhx6 and -8 act as transcription factors and coregulators of cell lineage specification. Known Lhx6 and -8 functions and targets are outlined in neurogenesis, craniofacial development, and germ cell differentiation. The underlying mechanisms of Lhx6 and -8 in regulating cell fate remain elusive. Whether Lhx6 and -8 affect functions in tissues and organs other than neural, craniofacial, oocytes, and germ cells is largely unexplored. Taken together, Lhx6 and -8 are important regulators of cell lineage specification and may act as one of the pivotal mediators of stem cell fate. Undoubtedly, future investigations of Lhx6 and -8 biology will continue to yield fascinating insights into tissue development and homeostasis, in addition to their putative roles in tissue regeneration and ageing.
Keywords: LIM homeobox genes; neurogenesis; palatogenesis; tooth development; transcriptional regulation.
© FASEB.
Figures
References
-
- McGinnis W., Krumlauf R. (1992) Homeobox genes and axial patterning. Cell 68, 283–302 - PubMed
-
- Boncinelli E. (1997) Homeobox genes and disease. Curr. Opin. Genet. Dev. 7, 331–337 - PubMed
-
- Hobert O., Westphal H. (2000) Functions of LIM-homeobox genes. Trends Genet. 16, 75–83 - PubMed
-
- Taira M., Evrard J. L., Steinmetz A., Dawid I. B. (1995) Classification of LIM proteins. Trends Genet. 11, 431–432 - PubMed
-
- Dawid I. B., Toyama R., Taira M. (1995) LIM domain proteins. C. R. Acad. Sci. III 318, 295–306 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

