Genetic screening reveals a link between Wnt signaling and antitubulin drugs
- PMID: 26149735
- PMCID: PMC4705004
- DOI: 10.1038/tpj.2015.50
Genetic screening reveals a link between Wnt signaling and antitubulin drugs
Abstract
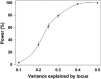
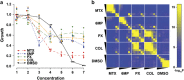
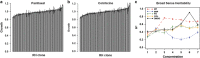
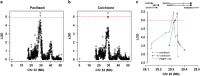
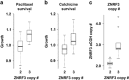
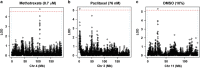
The antitubulin drugs, paclitaxel (PX) and colchicine (COL), inhibit cell growth and are therapeutically valuable. PX stabilizes microtubules, while COL promotes their depolymerization. But, the drug concentrations that alter tubulin polymerization are hundreds of times higher than their clinically useful levels. To map genetic targets for drug action at single-gene resolution, we used a human radiation hybrid panel. We identified loci that affected cell survival in the presence of five compounds of medical relevance. For PX and COL, the zinc and ring finger 3 (ZNRF3) gene dominated the genetic landscape at therapeutic concentrations. ZNRF3 encodes an R-spondin regulated receptor that inhibits Wingless/Int (Wnt) signaling. Overexpression of the ZNRF3 gene shielded cells from antitubulin drug action, while small interfering RNA knockdowns resulted in sensitization. Further a potent pharmacological inhibitor of Wnt signaling, Wnt-C59, protected cells from PX and COL. Our results suggest that the antitubulin drugs perturb microtubule dynamics, thereby influencing Wnt signaling.
Figures

References
-
- Collins FS, Green ED, Guttmacher AE, Guyer MS, US National Human Genome Research Institute. A vision for the future of genomics research. Nature 2003; 422: 835–847. - PubMed
-
- Fang Y. Ligand-receptor interaction platforms and their applications for drug discovery. Expert Opin Drug Discov 2012; 7: 969–988. - PubMed
-
- Pacholarz KJ, Garlish RA, Taylor RJ, Barran PE. Mass spectrometry based tools to investigate protein-ligand interactions for drug discovery. Chem Soc Rev 2012; 41: 4335–4355. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

