Generation of supercoils in nicked and gapped DNA drives DNA unknotting and postreplicative decatenation
- PMID: 26150424
- PMCID: PMC4551925
- DOI: 10.1093/nar/gkv683
Generation of supercoils in nicked and gapped DNA drives DNA unknotting and postreplicative decatenation
Abstract
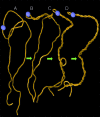
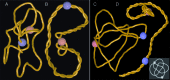
Due to the helical structure of DNA the process of DNA replication is topologically complex. Freshly replicated DNA molecules are catenated with each other and are frequently knotted. For proper functioning of DNA it is necessary to remove all of these entanglements. This is done by DNA topoisomerases that pass DNA segments through each other. However, it has been a riddle how DNA topoisomerases select the sites of their action. In highly crowded DNA in living cells random passages between contacting segments would only increase the extent of entanglement. Using molecular dynamics simulations we observed that in actively supercoiled DNA molecules the entanglements resulting from DNA knotting or catenation spontaneously approach sites of nicks and gaps in the DNA. Type I topoisomerases, that preferentially act at sites of nick and gaps, are thus naturally provided with DNA-DNA juxtapositions where a passage results in an error-free DNA unknotting or DNA decatenation.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
-
- Bates A.D., Maxwell A. DNA Topology. Oxford: Oxford University Press; 2005.
-
- Pulleyblank D.E. Of topo and Maxwell's dream. Science. 1997;277:648–649. - PubMed
-
- Rybenkov V.V., Ullsperger C., Vologodskii A.V., Cozzarelli N.R. Simplification of DNA topology below equilibrium values by type II topoisomerases. Science. 1997;277:690–693. - PubMed
-
- Yan J., Magnasco M.O., Marko J.F. A kinetic proofreading mechanism for disentanglement of DNA by topoisomerases. Nature. 1999;401:932–935. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

