Long noncoding RNA derived from CD244 signaling epigenetically controls CD8+ T-cell immune responses in tuberculosis infection
- PMID: 26150504
- PMCID: PMC4517270
- DOI: 10.1073/pnas.1501662112
Long noncoding RNA derived from CD244 signaling epigenetically controls CD8+ T-cell immune responses in tuberculosis infection
Abstract
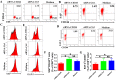
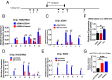
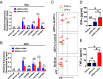
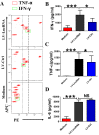
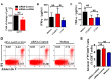
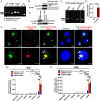
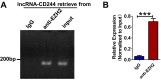
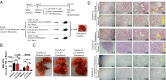
Molecular mechanisms for T-cell immune responses modulated by T cell-inhibitory molecules during tuberculosis (TB) infection remain unclear. Here, we show that active human TB infection up-regulates CD244 and CD244 signaling-associated molecules in CD8(+) T cells and that blockade of CD244 signaling enhances production of IFN-γ and TNF-α. CD244 expression/signaling in TB correlates with high levels of a long noncoding RNA (lncRNA)-BC050410 [named as lncRNA-AS-GSTT1(1-72) or lncRNA-CD244] in the CD244(+)CD8(+) T-cell subpopulation. CD244 signaling drives lncRNA-CD244 expression via sustaining a permissive chromatin state in the lncRNA-CD244 locus. By recruiting polycomb protein enhancer of zeste homolog 2 (EZH2) to infg/tnfa promoters, lncRNA-CD244 mediates H3K27 trimethylation at infg/tnfa loci toward repressive chromatin states and inhibits IFN-γ/TNF-α expression in CD8(+) T cells. Such inhibition can be reversed by knock down of lncRNA-CD244. Interestingly, adoptive transfer of lncRNA-CD244-depressed CD8(+) T cells to Mycobacterium tuberculosis (MTB)-infected mice reduced MTB infection and TB pathology compared with lncRNA-CD244-expressed controls. Thus, this work uncovers previously unidentified mechanisms in which T cell-inhibitory signaling and lncRNAs regulate T-cell responses and host defense against TB infection.
Keywords: CD8+ T cells; lncRNA; tuberculosis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures








Similar articles
-
Involvement of CD244 in regulating CD4+ T cell immunity in patients with active tuberculosis.PLoS One. 2013 Apr 30;8(4):e63261. doi: 10.1371/journal.pone.0063261. Print 2013. PLoS One. 2013. PMID: 23638187 Free PMC article.
-
The immunoregulatory role of CD244 in chronic hepatitis B infection and its inhibitory potential on virus-specific CD8+ T-cell function.Hepatology. 2010 Dec;52(6):1934-47. doi: 10.1002/hep.23936. Epub 2010 Nov 9. Hepatology. 2010. PMID: 21064032
-
Analysis of immune activation and clinical events in acute infectious mononucleosis.J Infect Dis. 2004 Jul 1;190(1):63-71. doi: 10.1086/421276. Epub 2004 Jun 8. J Infect Dis. 2004. PMID: 15195244
-
EZH2 as a Regulator of CD8+ T Cell Fate and Function.Front Immunol. 2020 Oct 6;11:593203. doi: 10.3389/fimmu.2020.593203. eCollection 2020. Front Immunol. 2020. PMID: 33117406 Free PMC article. Review.
-
Biochemical Methods To Investigate lncRNA and the Influence of lncRNA:Protein Complexes on Chromatin.Biochemistry. 2016 Mar 22;55(11):1615-30. doi: 10.1021/acs.biochem.5b01141. Epub 2016 Feb 24. Biochemistry. 2016. PMID: 26859437 Free PMC article. Review.
Cited by
-
Macrophage targeted iron oxide nanodecoys augment innate immunological and drug killings for more effective Mycobacterium Tuberculosis clearance.J Nanobiotechnology. 2023 Oct 10;21(1):369. doi: 10.1186/s12951-023-02103-x. J Nanobiotechnology. 2023. PMID: 37817142 Free PMC article.
-
Genome-wide target interactome profiling reveals a novel EEF1A1 epigenetic pathway for oncogenic lncRNA MALAT1 in breast cancer.Am J Cancer Res. 2019 Apr 1;9(4):714-729. eCollection 2019. Am J Cancer Res. 2019. PMID: 31105998 Free PMC article.
-
Host-Directed Targeting of LincRNA-MIR99AHG Suppresses Intracellular Growth of Mycobacterium tuberculosis.Nucleic Acid Ther. 2022 Oct;32(5):421-437. doi: 10.1089/nat.2022.0009. Epub 2022 Jul 27. Nucleic Acid Ther. 2022. PMID: 35895506 Free PMC article.
-
Transcriptional regulation of FOXP3 requires integrated activation of both promoter and CNS regions in tumor-induced CD8+ Treg cells.Sci Rep. 2017 May 9;7(1):1628. doi: 10.1038/s41598-017-01788-z. Sci Rep. 2017. PMID: 28487507 Free PMC article.
-
Long non-coding RNA transcripts in Mycobacterium tuberculosis-host interactions.Noncoding RNA Res. 2024 Dec 15;11:281-293. doi: 10.1016/j.ncrna.2024.12.005. eCollection 2025 Apr. Noncoding RNA Res. 2024. PMID: 39926616 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

